An Arabidopsis homolog of KIN17 physically interacts with the transcription factor SPL7 to prevent impaired plant growth during periods of limited copper availability.
Abstract
Proper copper (Cu) homeostasis is required by living organisms to maintain essential cellular functions. In the model plant Arabidopsis (Arabidopsis thaliana), the SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE7 (SPL7) transcription factor participates in reprogramming global gene expression during Cu insufficiency in order to improve the metal uptake and prioritize its distribution to Cu proteins of major importance. As a consequence, spl7 null mutants show morphological and physiological disorders during Cu-limited growth, resulting in lower fresh weight, reduced root elongation, and chlorosis. On the other hand, the Arabidopsis KIN17 homolog belongs to a well-conserved family of essential eukaryotic nuclear proteins known to be stress activated and involved in DNA and possibly RNA metabolism in mammals. In the study presented here, we uncovered that Arabidopsis KIN17 participates in promoting the Cu deficiency response by means of a direct interaction with SPL7. Moreover, the double mutant kin17-1 spl7-2 displays an enhanced Cu-dependent phenotype involving growth arrest, oxidative stress, floral bud abortion, and pollen inviability. Taken together, the data presented here provide evidence for SPL7 and KIN17 protein interaction as a point of convergence in response to both Cu deficiency and oxidative stress.
Copper (Cu) is an essential micronutrient for all living organisms as a cofactor of many proteins involved in fundamental biochemical processes. In plants, for instance, the redox properties of Cu are used by the cytochrome c oxidase or the Cu-zinc (Zn) superoxide dismutases (SODs) CSD1 and CSD2 to perform the terminal oxidation in the mitochondrial electron chain or the superoxide anion dismutation, respectively. In addition, the most abundant Cu protein, plastocyanin (PC), contributes to the electron transfer between the two chloroplastidial photosystems (Puig et al., 2007; Burkhead et al., 2009). Furthermore, Cu homeostasis in plants is receiving increasing interest since Cu plays a structural role in the ethylene receptors, the molybdenum cofactor, or the salicylic acid receptor NONEXPRESSER OF PR GENES1 (Rodríguez et al., 1999; Kuper et al., 2004; Wu et al., 2012). However, an excess of free Cu disturbs cellular components due to the peroxidation of lipids, changes in the tertiary structure of proteins, metal substitution at the active centers of particular enzymes, and mutations in nucleic acids (Halliwell and Gutteridge, 1984).
Consequently, a fine-tuned homeostatic network has evolved in order to keep Cu levels within the proper range. Although principal components of the Cu homeostatic network seem to be conserved in all eukaryotes, the main evolutionary divergences are found in the regulatory mechanisms (Puig et al., 2007; Burkhead et al., 2009). Indeed, plants possess a unique family of transcription factors, known as SQUAMOSA PROMOTER-BINDING PROTEINS (SBPs), sharing a conserved DNA-binding domain known as the SBP domain. From single-celled algae like Chlamydomonas reinhardtii to the eudicot Arabidopsis (Arabidopsis thaliana), a SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE (SPL) transcription factor orchestrates major elements of the Cu deficiency response (Kropat et al., 2005; Yamasaki et al., 2009; Bernal et al., 2012). Like the Cu response regulator CRR1 in C. reinhardtii, Arabidopsis SPL7 directly binds to a GTAC motif of Cu response elements within the promoter regions of Cu-responsive genes (Birkenbihl et al., 2005; Kropat et al., 2005; Yamasaki et al., 2009). A recent comparative transcriptome profiling reinforced the key role that SPL7 plays in reprogramming global gene expression in order to promote Cu uptake and economization during Cu-limiting periods (Bernal et al., 2012). In fact, SPL7 induces a high-affinity Cu uptake system based on the newly identified plasma membrane metalloreductases FERRIC REDUCTION OXIDASE4 and FERRIC REDUCTION OXIDASE5 and the plasma membrane-related Cu transport proteins COPPER TRANSPORTER1 (COPT1), COPT2, and COPT6 (Bernal et al., 2012; Jung et al., 2012; Garcia-Molina et al., 2013; Perea-García et al., 2013). Simultaneously, SPL7 triggers the so-called Cu economy mode by inducing the iron (Fe) SOD FSD1 and a set of microRNAs (denoted Cu-microRNAs) to down-regulate nonessential Cu proteins, such as miR398, targeting CSD1 and CSD2, or miR408, repressing plantacyanin and laccases (Abdel-Ghany and Pilon, 2008). Therefore, the coordinated substitution of the SODs at the chloroplast allows the Cu reallocation toward PC (Burkhead et al., 2009; Yamasaki et al., 2009; Bernal et al., 2012).
How Arabidopsis senses Cu deficiency remains a largely unanswered question. Intriguingly, SPL7 transcripts are detected in all plant parts, and their levels remain largely constant regardless of Cu availability, suggesting posttranscriptional regulation (Yamasaki et al., 2009; Bernal et al., 2012). Cu2+ ions may negatively interfere with the DNA-binding activity of the SPL7 transcription factor in vitro, as proposed by Sommer and coworkers (2010), but how Cu is delivered to SPL7 remains unknown. Also, to enable the modulation of SPL7 activity in different organs and tissues, it is not unlikely that SPL7 interacts with different proteins. Based on this assumption, the identification of proteins able to associate with SPL7 would contribute to a better understanding of its function, shed light on the inputs that govern Cu homeostasis, and reveal new roles for Cu in plant development.
In this work, we addressed this question by providing supporting evidence for a physical interaction in planta between SPL7 and a KIN17-like protein. The KIN17 protein family constitutes a group of extremely well-conserved eukaryotic proteins that bind preferentially to bent DNA (Mazin et al., 1994). KIN17 homologs in mammals have been reported to participate in DNA replication, cell growth progression, and the proliferation and preserving of DNA integrity under detrimental conditions (Kannouche et al., 1998; Biard et al., 2002; Masson et al., 2003). To date, there are no available data concerning the function of KIN17 in plants. Here, we focused on the protein coded by the Arabidopsis AT1G55460 gene, which shows the highest degree of identity to mammalian orthologs. We uncovered a new function for the Arabidopsis KIN17 protein in promoting Cu-responsive genes by means of a physical interaction with SPL7. Moreover, the double mutant spl7-2 kin17-1 reveals the convergence of SPL7 and KIN17 in a common pathway to alleviate oxidative stress and guarantee proper growth and fertility during Cu-limiting periods.
RESULTS
KIN17 Interacts with SPL7 in Vivo
Given that SPL7 (AT5G18830) is largely constitutively expressed throughout the plant regardless of Cu availability, it is suggested that at least part of its regulation takes place posttranslationally (Yamasaki et al., 2009; Bernal et al., 2012). In order to identify putative SPL7-interacting proteins that may be involved in such a regulation, a yeast two-hybrid (Y2H) screening was conducted. Using as bait an SPL7 fragment encompassing the SBP domain and the IRPGC signature (amino acid residues 133–762), a prey corresponding to the first 151 amino acid residues of the protein encoded by AT1G55460 was identified (Supplemental Fig. S1). The isolated candidate represents a member of an extremely well-conserved protein family in Eukaryota containing (1) a C2H2-like zinc finger domain (IPR015880; Mazin et al., 1994), (2) a so-called Kin17 DNA/RNA-binding domain (IPR019447), and (3) a Kyrpides, Ouzounis, Woese (KOW) domain (IPR005824; Kyrpides et al., 1996; Supplemental Figs. S1 and S2). Although the orthologs of this protein in mammals have been described to participate in DNA replication and repair, its role in Arabidopsis or other plants is so far unknown. Nevertheless, we will refer to this protein hereinafter as KIN17.
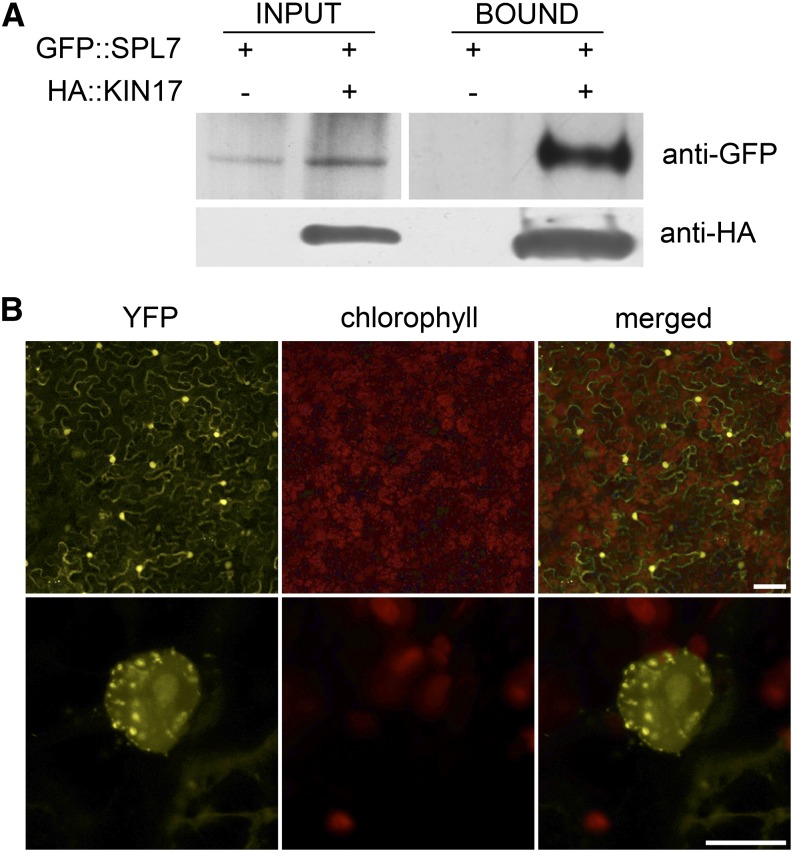
To confirm the interaction between SPL7 and KIN17 in planta, we first carried out an in vivo coimmunoprecipitation assay. Thereto, protein extracts from tobacco (Nicotiana benthamiana) leaves transiently coexpressing hemagglutinin (HA)-tagged KIN17 (HA::KIN17) and GFP-tagged SPL7 (GFP::SPL7) recombinant fusion proteins were prepared. Subsequent western-blot analysis of a pull-down fraction obtained using a specific antibody to the HA epitope confirmed the coimmunoprecipitation of GFP::SPL7 (Fig. 1A). In addition, the association SPL7-KIN17 was also addressed by a bimolecular fluorescence complementation (BiFC) strategy based on the reconstitution of yellow fluorescent protein (YFP) after translational fusion of its N-terminal or C-terminal half to SPL7 or KIN17 (nYFP::SPL7 or cYFP::KIN17), respectively, as described in “Materials and Methods.” Whereas only background fluorescence could be observed when transiently expressing the individual constructs in tobacco leaves, their coinfiltration regenerated a specific YFP signal in nuclei with a speckled pattern (Fig. 1B; Supplemental Fig. S3).
Figure 1.
KIN17 physically interacts with SPL7. A, KIN17 coimmunoprecipitates with SPL7. Total proteins from tobacco leaves transiently expressing the indicated tagged proteins were extracted (input) as described in “Materials and Methods” and pulled down by means of an anti-HA antibody (bound). Input and bound fractions were then assayed by western blot with anti-GFP-HRP antibodies to test for SPL7 coimmunoprecipitation. Membrane reprobed with an anti-HA-HRP is shown as a control for the pull down. B, BiFC assay for the nYFP::SPL7 and cYFP::KIN17 interaction. Epidermal cells of coinfiltrated tobacco leaves were examined after 3 d for restoration of YFP fluorescence using laser confocal microscopy. The YFP and chlorophyll signals are shown in color-separated and merged images. Bars = 100 µm (top row) and 10 µm (bottom row).
Together, these results corroborate that KIN17 is indeed able to physically interact with SPL7 in planta. Moreover, based on the fragment originally identified in the Y2H assay, we propose that the domain responsible for the interaction would be located within the first 151 amino acids of KIN17.
KIN17 Is a Nuclear Protein Mainly Expressed in Aerial Tissues and Pollen
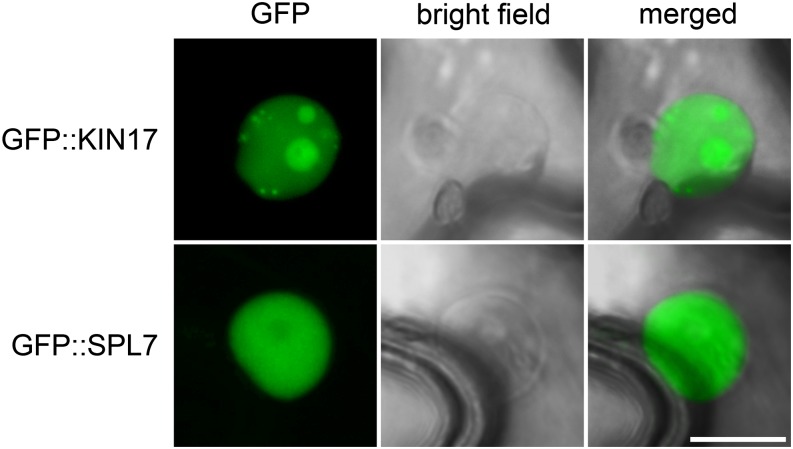
Once we validated that KIN17 and SPL7 are prone to interact with each other, we investigated the possibility of such an interaction occurring in planta by comparing their subcellular localization and expression patterns. Both SPL7 and KIN17 contain bipartite nuclear localization signals and are thus predicted to be targeted to the nucleus (Kannouche et al., 1997; Sommer et al., 2010). Confocal microscopy of tobacco leaf epidermal cells transiently expressing N-terminal GFP-tagged versions of KIN17 (GFP::KIN17) and SPL7 (GFP::SPL7) confirmed an exclusive nuclear localization. However, whereas GFP::KIN17 showed a mottled distribution in nuclei and illuminated nucleoli, GFP::SPL7 labeled the nucleoplasm homogenously, but not or less fully the nucleolus (Fig. 2). Taking this observation into consideration with our previous BiFC experiments, we conclude that KIN17 and SPL7 colocalize within nuclei, where their interaction may be associated with nuclear foci.
Figure 2.
KIN17 and SPL7 proteins localize to the nucleus in tobacco leaves. The entire KIN17 and SPL7 coding sequences were N-terminal fused to GFP and transiently expressed in tobacco leaves. Samples were observed using laser confocal microscopy 3 d post infiltration. The GFP fluorescence signal and the transmitted light (bright field) of representative samples are shown in separated and merged images. Bar = 10 µm.
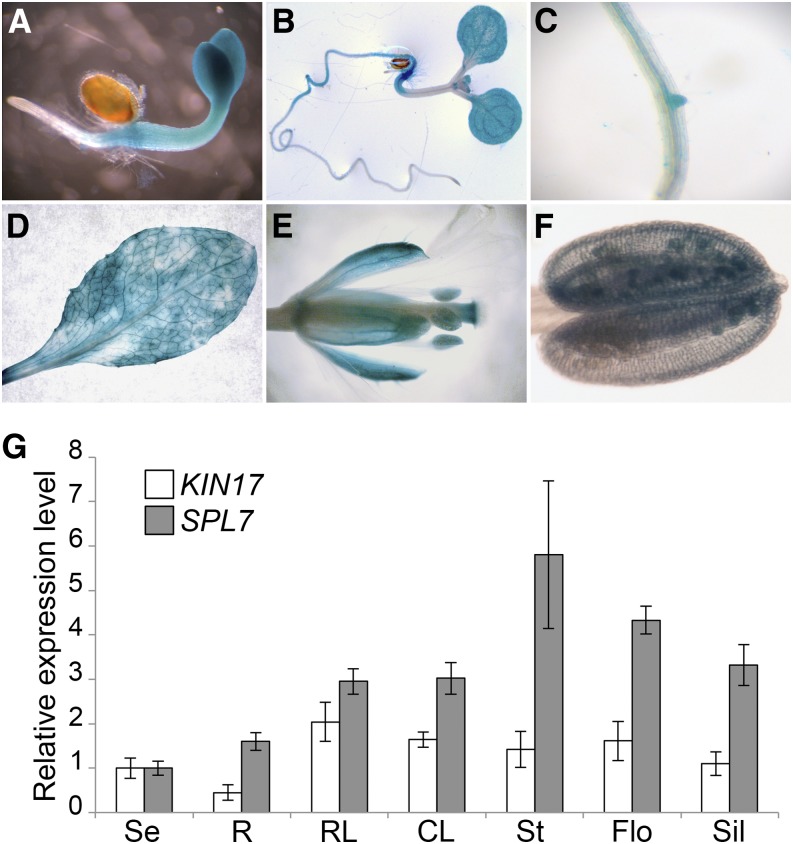
Next, in order to obtain a detailed picture of the spatial and temporal distribution of KIN17 in planta, stable transgenic lines expressing a GUS reporter gene under the control of the KIN17 promoter region to 673 bp upstream of its start codon (pKIN17::GUS) were generated. Early GUS expression became detectable in cotyledons, hypocotyls, and the vicinity of the radicle in 4-d-old seedlings grown on one-half-strength Murashige and Skoog (1/2 MS) medium (Fig. 3A). During subsequent development, GUS staining disappeared from the cotyledon petioles and hypocotyl except for its distal region, where it continued into the main root. Emerging leaflets and lateral roots also developed staining as well as the root vasculature (Fig. 3, B and C). The distal part of the root always remained unstained (Fig. 3, A and B). Adult plants showed a more or less homogenous GUS signal in rosette and cauline leaves as well as in the sepals of flowers (Fig. 3, D and E). Interestingly, in addition to the stylar region of the pistil, mature pollen grains developed a positive GUS staining (Fig. 3, E and F). In addition, to confirm our data based on GUS expression, the KIN17 mRNA levels were determined in whole seedlings and selected organs of adult plants by quantitative PCR (qPCR). Among the assayed organs, KIN17 transcripts were mainly detected at comparable levels in rosette and cauline leaves, stems, and inflorescences, whereas seedlings, siliques, and, especially, roots displayed relatively more moderate expression levels (Fig. 3G). For comparison, SPL7 transcript levels were determined in the same samples and found to be broadly expressed as well. Under our plant growth conditions, SPL7 transcript levels were highest in the stems, inflorescences, and siliques and lowest in seedlings and roots (Fig. 3G). Taken together, our data clearly point to a partial overlapping expression and subcellular localization of KIN17 and SPL7, thus providing a prerequisite for a functional interaction between the two proteins.
Figure 3.
Characterization of the KIN17 spatial expression pattern. A to F, Histochemical GUS staining in pKIN17::GUS lines. A and B, Five-day-old (A) and 7-d-old (B) seedlings. C, Root vasculature. D, Rosette leaves. E, Postanthesis flowers. F, Anthers. G, KIN17 and SPL7 distribute mainly on aerial organs. Total RNA from seedlings and different adult plant organs, seedlings (Se), roots (R), rosette leaves (RL), cauline leaves (CL), stems (St), flowers (Flo), and siliques (Sil), was isolated and reverse transcribed to cDNA. KIN17 and SPL7 mRNA relative amounts were measured by qPCR with specific oligonucleotides and normalized to ACTIN1 and ELONGATION FACTOR1α levels. Error bars correspond to the sd of technical replicates (n = 3).
KIN17 Promotes the Expression of SPL7 Targets
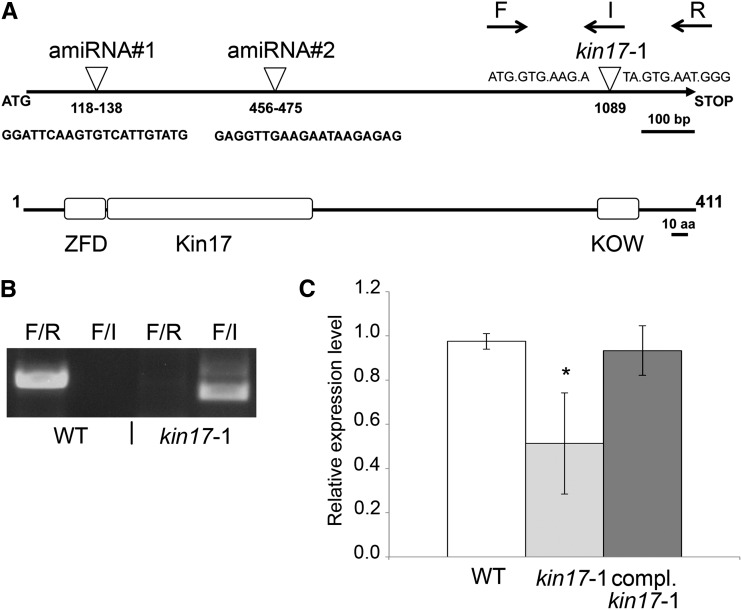
To evaluate how KIN17 contributes to Arabidopsis metabolism, we obtained line N677041 from the SALK transfer DNA (T-DNA) collection (hereinafter kin17-1) and confirmed the insertion of a T-DNA within the region encoding the KOW motif at 1,089 nucleotides downstream of the translational start site (Fig. 4, A and B). Homozygous kin17-1 individuals were selected for further analysis, but no obvious phenotypic abnormalities were noticed when grown on soil or in vitro on medium containing different Cu concentrations. Based on an approximately 50% reduction in KIN17 mRNA levels in 7-d-old mutant seedlings (Fig. 4C), together with the position of the T-DNA, kin17-1 could represent an allele version with residual function.
Figure 4.
Isolation of the kin17-1 mutant line. A, Scheme representing the KIN17 gene and the translated protein with its conserved domains defined. The T-DNA insertion in the kin17-1 line is indicated by an inverted triangle and corresponding flanking sequences. The positions of the oligonucleotides used to genotype the lines are shown by arrows (F, I, and R). The Zn finger domain (ZFD), Kin17, and KOW domains are indicated with squares. aa, Amino acids. B, Discrimination of homozygous kin17-1 plants. kin17-1 plants were self-crossed, and the offspring was genotyped with the oligonucleotide combinations indicated in A: for the wild-type (WT) allele, F + R; for the insertion allele, F + I. C, The kin17-1 line is a knockdown mutant. Total RNA from 6-d-old seedlings of the wild type, kin17-1, and pKIN17::KIN17 in the kin17-1 mutant background grown on 1/2 MS medium supplemented with Suc and BCS was prepared and reverse transcribed to cDNA. KIN17 mRNA relative levels were determined by qPCR and normalized to ACTIN1 and ELONGATION FACTOR1α levels. Error bars correspond to sd values of at least three independent biological replicates. The asterisk indicates a statistically significant difference from the wild type (P < 0.05).
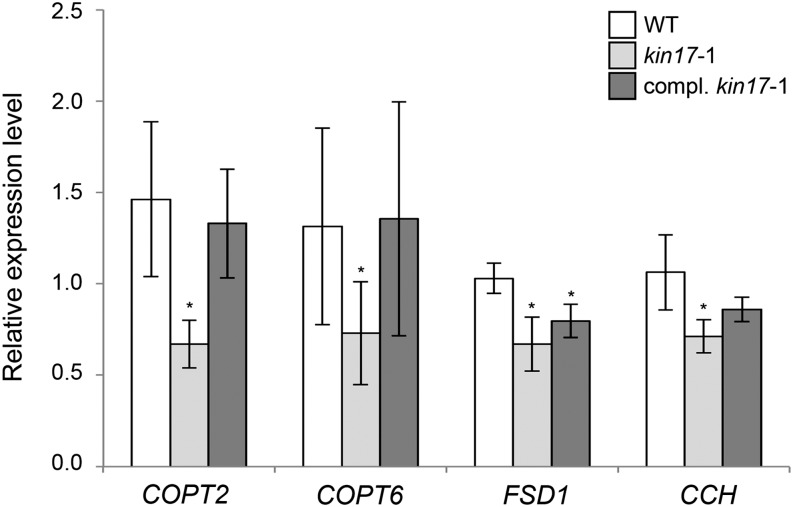
Our data supported an interaction between KIN17 and SPL7 in aerial tissues. Thus, to better understand the purpose of this interaction, relative mRNA levels of selected shoot-expressed SPL7 targets, such as the Cu high-affinity transport proteins encoding COPT2 and COPT6 genes, FSD1, and the metallochaperone encoding COPPER CHAPERONE (CCH) gene (Himelblau et al., 1998; Mira et al., 2001; Garcia-Molina et al., 2013; Perea-García et al., 2013), were compared between the kin17-1 mutant and the wild type. Strikingly, COPT2 and COPT6 transcript levels appeared to be approximately 50% reduced, whereas FSD1 and CCH levels were reduced by approximately 35% in 7-d-old kin17-1 seedlings grown on Cu-deficient medium (Fig. 5). Moreover, the expression of a KIN17 transgene under the control of its own promoter (pKIN17::KIN17) in the kin17-1 background not only restored KIN17 expression to wild-type levels but also those of the selected SPL7 targets to a certain extent (Figs. 4C and 5). Hence, we concluded that the observed altered expression of the selected genes is primarily KIN17 dependent and supports the hypothesis that KIN17 interacts with SPL7 in tuning the Cu deficiency response.
Figure 5.
Deregulation of selected SPL7 targets in the kin17-1 mutant. Total RNA from 7-d-old seedlings of the wild type (WT), kin17-1, and pKIN17::KIN17 in the kin17-1 mutant background grown on 1/2 MS medium supplemented with Suc and 50 µm BCS was prepared and reverse transcribed to cDNA. The relative mRNA levels of the indicated targets were determined by qPCR and normalized to ACTIN1 and ELONGATION FACTOR1α transcripts. Error bars correspond to sd values of at least three independent biological replicates. Asterisks indicate statistically significant differences from the wild type (P < 0.05).
The spl7 Mutant Cu Deprivation Response Is Further Impeded by the kin17 Mutation
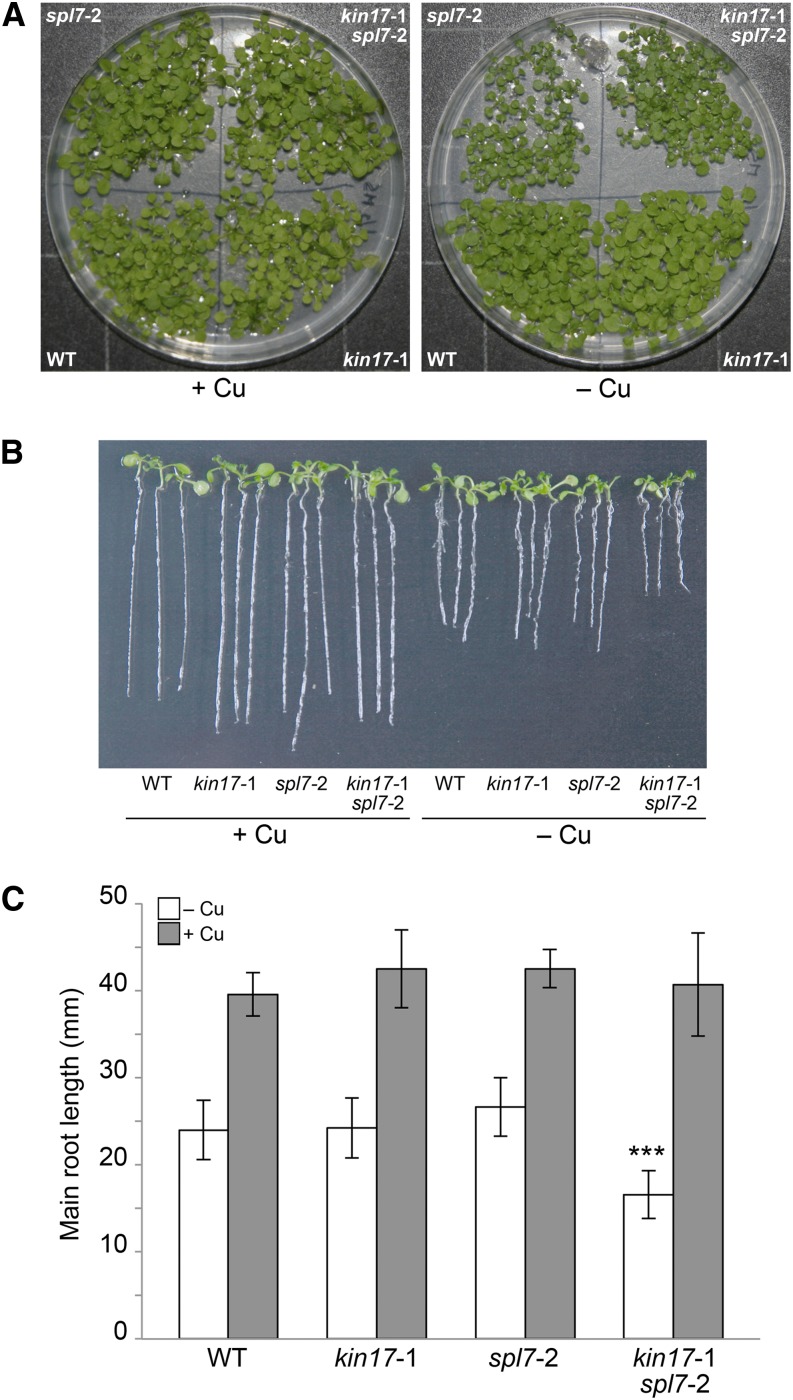
To gain insight into the genetic relationship between KIN17 and SPL7, the single mutants kin17-1 and spl7-2 (Bernal et al., 2012) were crossed, and the F2 offspring were genotyped in order to isolate a double homozygous mutant (Supplemental Fig. S4A). During the initial stages, we observed a slight growth delay in both the spl7-2 single and kin17-1 spl7-2 double mutants, in comparison with the wild-type and kin17-1 lines, when grown on 1/2 MS medium supplemented with the extracellular Cu+-specific chelator bathocuproine disulfonate (BCS; Fig. 6A). To further quantify this phenomenon, we measured the main root length of 6-d-old seedlings grown on vertical plates. On Cu-supplemented medium, the four assayed lines showed similar root length. However, whereas Cu deprivation inhibited root growth to around 60% in the wild type and the single mutants, the root length of the double mutant kin17-1 spl7-2 was reduced to 40% in comparison with that in Cu-replete medium, stressing its enhanced susceptibility to Cu scarcity (Fig. 6, B and C).
Figure 6.
Double mutant kin17-1 spl7-2 seedlings exhibit hypersensitivity to Cu deprivation. A, Ten-day-old seedlings of the wild type (WT), kin17-1, spl7-2, and kin17-1 spl7-2 germinated and grown on 1/2 MS supplemented with Suc and 5 µm CuSO4 (+Cu) or 50 µm BCS (–Cu). B, The same four lines were grown on vertical plates under identical conditions for 6 d. Three representative seedlings of each line and treatment were photographed. C, Measurement of main root lengths of seedlings as grown and shown in B. Error bars indicate sd (n ≥ 10 biological replicates). Asterisks indicate a statistically significant difference from the wild type (P < 0.001).
To examine later stages of development, we performed hydroponic culturing during which both the spl7-2 and the kin17-1 spl7-2 lines manifested obvious Cu-dependent impairments in growth. This culturing method, however, had a more dramatic impact on the double mutant (Supplemental Fig. S4B) and made us decide to alleviate growing conditions by cultivating on standard soil with a very mild Cu deficiency (hereinafter denoted as –Cu). As stated previously, the kin17-1 mutant did not show a differential phenotype under these conditions, whereas the spl7-2 mutant exhibited reductions in size and number of siliques (Fig. 7; Bernal et al., 2012). Intriguingly, the double mutant kin17-1 spl7-2 showed a more severely affected phenotype, consisting of stunted growth with a purple-colored body, altered inflorescence architecture due to early growth arrest of the main shoot and loss of apical dominance, deteriorated flowers, and reduced silique production (Fig. 7, A and C–E). All phenotypic aspects were clearly Cu dependent, as deduced from their restoration when applying extra Cu to the soil (+Cu; Fig. 7B). Taken together, our observations suggest that the concerted action between KIN17 and SPL7 is necessary to achieve proper growth and development under Cu-limiting conditions.
Figure 7.
Phenotypes of the double mutant kin17-1 spl7-2 grown on soil. A and B, Representative 6-week-old wild-type (WT), kin17-1, spl7-2, and kin17-1 spl7-2 double mutant plants cultivated under long-day conditions on standard soil (A) or on soil treated with CuSO4 (+Cu; B). C to E, Detailed macro images of inflorescences of selected kin17-1 spl7-2 plants grown on non-Cu-supplemented soil as shown in A.
KIN17 and SPL7 Contribute to Alleviate Oxidative Stress under Cu Limitation
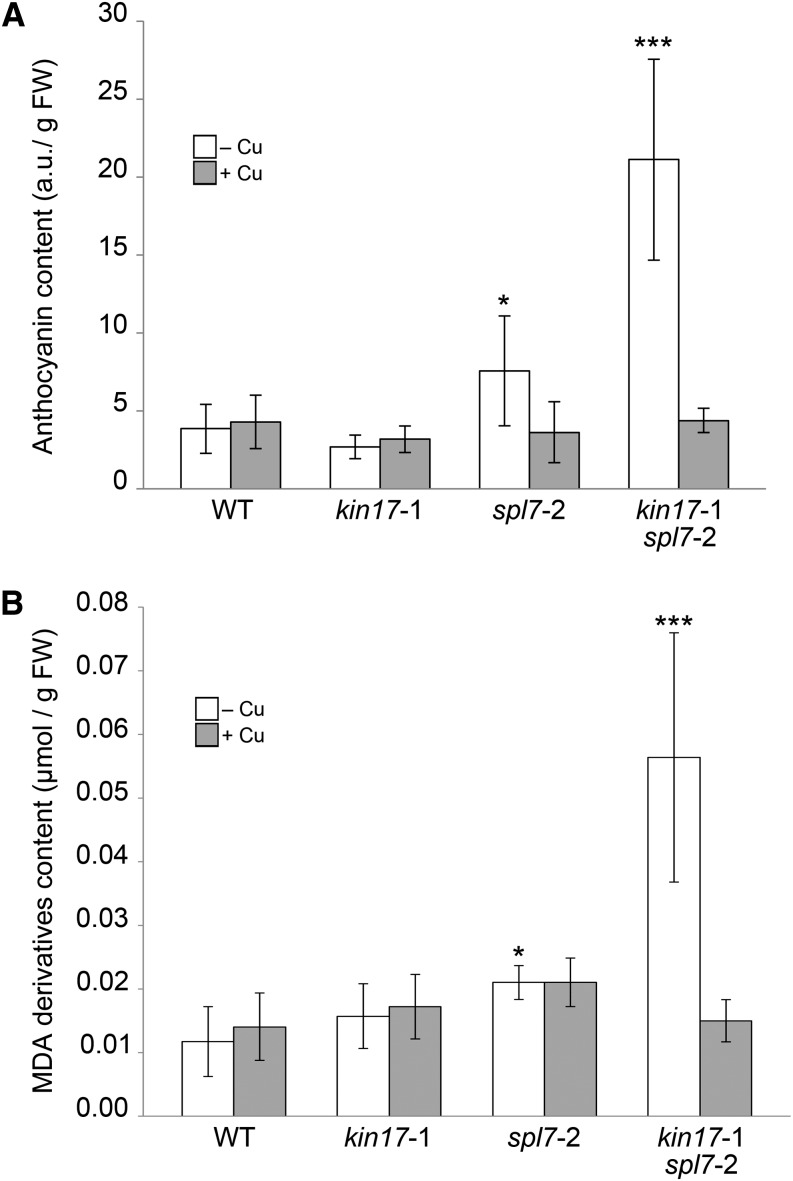
To trace possible causes of the augmented Cu-deficiency phenotype in the double mutant, we paid attention to the purple pigmentation, in particular of the leaf abaxial sides of kin17-1 spl7-2 plants, which could be a consequence of oxidative stress (Supplemental Fig. S4C). Therefore, the anthocyanin content and the degree of lipid peroxidation of rosette leaves were determined to infer oxidative stress. Whereas leaves of wild-type and kin17-1 plants grown on –Cu soil contained a basal amount of anthocyanin, the pigment levels in those of the spl7-2 mutant and the double mutant were 1.5- and 5-fold increased, respectively (Fig. 8A). Similarly, a significant increment in the amount of malondialdehyde derivatives (i.e. the end products of lipid peroxidation) could be observed in leaves of spl7-2 (2-fold) and the double mutant (5-fold) when grown on –Cu soils, suggesting a higher rate of lipid peroxidation (Fig. 8B). Again, the basal levels for both parameters were recovered by Cu application to the soil (Fig. 8). Based on these data, we suggest that the convergent action of KIN17 and SPL7 is required to optimally counteract the effects of oxidative stress under Cu-deprived conditions in order to maintain proper growth and development.
Figure 8.
Measurement of oxidative stress-related parameters. A, The anthocyanin content is increased in kin17-1 spl7-2 double mutants when grown on standard soils. Anthocyanins from 1-month-old rosette leaves were extracted and quantified as described in “Materials and Methods” and normalized against fresh weight (FW). B, Lipid peroxidation is increased in kin17-1 spl7-2 grown on standard soils. Malondialdehyde (MDA) amounts in 1-month-old rosette leaves from the lines indicated were determined as described in “Materials and Methods” and normalized against fresh weight. Error bars correspond to the sd of at least six independent biological samples. Asterisks indicate statistically significant differences from the wild type (WT; *P < 0.05, ***P < 0.001).
KIN17 and SPL7 Ensure Qualitatively and Quantitatively the Production of Pollen under Cu Limitation
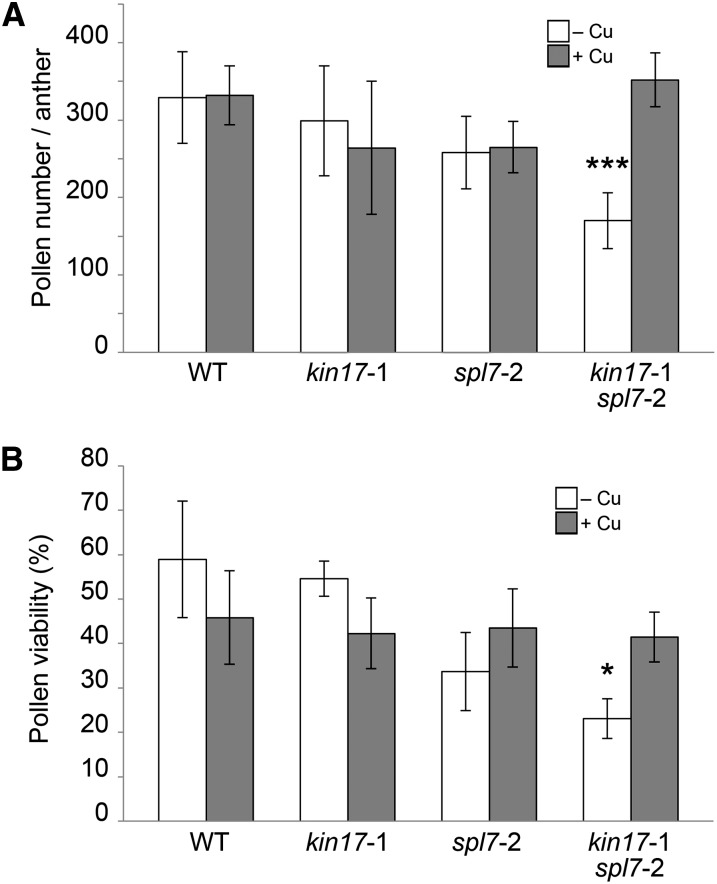
The strong reduction in fertility is one of the most dramatic aspects of the kin17-1 spl7-2 double mutant phenotype. Cu deprivation is known to have a negative impact on pollen development and viability (Dell, 1981; Sancenón et al., 2004). As pKIN17 expression could be detected at this level (Fig. 3F), we investigated to what extent the semisterility of the double mutant could be due to KIN17 malfunction in pollen. Thereto, anthers were dissected from the fifth and the 10th flowers formed in the main inflorescence, and the average amount of pollen per anther was determined. Wild-type flowers showed equal amounts of pollen per anther when grown on –Cu or +Cu soils. For the kin17-1 and spl7-2 single mutant lines, similar results were obtained, albeit with an overall slight decrease in pollen number to approximately 80% of the wild-type value (Fig. 9A). Interestingly, however, the double mutant showed a more significant reduction to approximately 50% of the wild-type value when grown on –Cu soil (Fig. 9A). Moreover, it should be noted that the pollen production of the double mutant was actually more dramatically affected, as a considerable amount of its flowers ceased development prematurely, rendering them useless for dissection of their anthers (Fig. 7, compare A and C–E). Importantly, supplying Cu to the soil restored the amount of pollen to normal (Fig. 9A).
Figure 9.
Determination of pollen production and viability. A, Pollen production in kin17-1 spl7-2 double mutant anthers is reduced compared with the wild type (WT) when grown on standard soil. Pollen was extracted from pooled anthers dissected from flowers just before anthesis in positions 5 to 10 of main inflorescences of plants cultivated as shown in Figure 7. Error bars indicate sd values of six countings representing 30 anthers in total per genotype. B, Impact of different Cu regimes on pollen viability. Pollen from recently opened flowers was immersed in a drop of FDA-Suc solution, and the percentage of fluorescent pollen was scored. Error bars represent sd values of at least four independent biological experiments. Asterisks indicate statistically significant differences in comparison with the wild type (*P < 0.05, ***P < 0.001).
Next, we investigated pollen viability by means of the fluorescein diacetate (FDA) test. By this test, only viable pollen is able to internalize and metabolize FDA and thereby become fluorescent. According to this test and when grown on –Cu soil, the viability of spl7-2 mutant pollen suffered an approximately 45% decrease compared with the viability of wild-type and kin17-1 pollen, whereas the combination of both single mutants resulted in a reduction of approximately 60% (Fig. 9B). Interestingly, Cu treatment restored the stated impairments to wild-type levels, although an approximately 25% decrease could be detected in all four lines assayed in comparison with the wild-type values under –Cu. Furthermore, there were no significant differences in the germination rates of harvested seeds among lines and treatments (data not shown). Taken together, these data indicate that both reduced quantity and quality of pollen strongly contribute to the semisterile phenotype of the double mutant kin17-1 spl7-2. Consequently, these data constitute new evidence concerning the pollen sensitivity to both Cu deprivation and excess.
DISCUSSION
In this study, we provide experimental evidence based on three independent in vivo approaches that SPL7 can physically interact with a previously functionally unknown KIN17 protein in Arabidopsis.
Two main studies aimed at understanding the SPL7 function in Arabidopsis Cu homeostasis concluded that SPL7 transcript levels are largely independent of Cu availability in the medium (Yamasaki et al., 2009; Bernal et al., 2012). Similar results had earlier been obtained with respect to the expression of the C. reinhardtii SPL7 ortholog, CRR1 (Kropat et al., 2005). These observations support the idea that SPL7 function is modulated posttranscriptionally and/or posttranslationally. For instance, Sommer and coworkers (2010) showed how Cu2+ ions may negatively interfere with the DNA-binding activity of both the SPL7 SBP domain and C. reinhardtii CRR1, thereby revealing a possible fundamental mechanism in SBP transcription factor-dependent Cu homeostasis. However, Cu ions are not believed to move freely within cells (Rae et al., 1999), and how Cu ions would be delivered to SPL7 remains unknown. Furthermore, higher plants like Arabidopsis are expected to display differential SPL7-dependent tissue/organ responses, at least with respect to uptake, distribution, and demands. Taken together, such considerations suggest possible tissue-specific protein interactions likely to play important roles in modulating SPL7 function.
In combining pKIN17::GUS transgenic reporter line data and qPCR analysis of multiple tissues and organs, we demonstrated that both the SPL7- and KIN17-related genes find extensive overlap in their temporal and spatial expression domains. Moreover, both Arabidopsis KIN17 and SPL7 contain predicted bipartite nuclear localization signals. Indeed, both SPL7 and KIN17 proteins were allocated to the nucleus when expressed in heterologous tobacco leaves, but intranuclear distribution differed. Whereas GFP-tagged SPL7 protein homogenously stained the nucleoplasm but seemed to be excluded from the nucleolus, GFP-tagged KIN17 exhibited a preferential nucleolar localization in combination with a dot-like distribution within the rest of the nucleus.
That SPL7 and KIN17 do indeed physically interact within nuclei can be deduced from the restoration of the YFP signal in our BiFC experiments that, in concert with the above-described observations, mimicked the KIN17 distribution pattern. We thus conclude that the SPL7-KIN17 interaction should take place within subnuclear foci or speckles.
Interestingly, a highly similar speckled nuclear localization pattern has also been reported for the mammalian KIN17 orthologs and related to their capacity to bind curved DNA (Kannouche et al., 1997). Consequently, the discrete distribution of bent DNA within nuclei would precede the above-described pattern. Indeed, ultrastructural assays in cultured human cells colocalized the human KIN17 with newly synthesized DNA labeled with bromodeoxyuridine as well as with DNA replication-associated markers, such as the proliferating cell nuclear antigen, the replication protein A, or the DNA polymerase-α (Biard et al., 2003; Miccoli et al., 2005). These observations point out that KIN17 is associated with the nonchromatin nuclear structures named replication foci. In agreement with this fact, truncations affecting the conserved domains in mouse KIN17 lead to impairments in DNA binding and, thus, a partial mislocalization of the protein (Kannouche et al., 1997).
Because replication origins and illegitimate recombination junctions constitute major sites for bent DNA, most of the studies in mammals have been focused on the role of KIN17 in DNA replication and the maintenance of genomic integrity in response to genotoxic agents or ionizing radiation (Angulo et al., 1989; Kannouche et al., 1998; Biard et al., 2002, 2003; Masson et al., 2003). However, bent or curved DNA sequences in upstream promoter regions could also constitute a preferred conformation for the binding of transcription factors (Ohyama, 2005). Hence, we propose a novel function for KIN17 in accompanying transcription factors in order to facilitate and ameliorate the affinity for their cis-elements.
In support of such a function, the partial KIN17 suppression in the kin17-1 T-DNA line relatively abrogates the mRNA levels of selected SPL7 targets under Cu-starving periods. Given that KIN17 is not expressed in the main part of the root, we propose that KIN17 primarily interacts with SPL7 to specifically enhance SPL7 activity in the aerial parts of the plant. Based on this assumption, the remaining mRNA levels of the here-analyzed Cu-responsive genes could be attributed either to their expression in roots or to an SPL7-independent activation. In fact, COPT2 and COPT6 may reflect such transcriptional behavior. COPT2 has been reported recently to be expressed during Cu-deficient periods in the shoot, but also in the differentiation zone of the root, in a nonoverlapping manner to KIN17 (Perea-García et al., 2013). In the case of COPT6, although its expression is mostly confined to aerial tissues, an SPL7-independent regulation maintains a basal expression, which would correspond to the remaining levels in kin17-1 (Jung et al., 2012; Garcia-Molina et al., 2013). Finally, based on the observed minimal repression for FSD1 and CCH, we have to conclude that not all the SPL7 targets are promoted by KIN17 in aerial plant parts. This category of Cu-responsive genes is triggered either by SPL7 alone or in concert with additional as yet unidentified SPL7-interacting proteins.
However, a more global role for KIN17 in Arabidopsis development independent of SPL7 cannot be ruled out. In fact, KIN17-like proteins are found in all eukaryotes (le Maire et al., 2006), whereas SBP-domain proteins like SPL7 seem plant specific (Yamasaki et al., 2008). Furthermore, KIN17 orthologs in higher eukaryotes display a high degree of sequence similarity (Carlier et al., 2007). Consequently, it is tempting to speculate that the Arabidopsis KIN17 preserved functions similar to those described for mammals, such as in DNA replication and repair (Biard et al., 2002; Masson et al., 2003; Miccoli et al., 2005). Also, the enhanced growth defects and signs of oxidative stress shown by the kin17-1 spl7-2 double mutant grown on standard soil in comparison with the spl7-2 single mutant could indicate a more general role of KIN17 in abiotic stress responses. From this perspective, KIN17 may play a previously unrecognized important role in plant growth and development that deserves a more comprehensive study.
Despite the current lack of knowledge on the function of KIN17 in plants, we considered the fact that KIN17 enhances SPL7 activities as a chance to explore new functions for Cu in plant homeostasis. Indeed, assuming a conserved role of KIN17, one could hypothesize that Cu is required to face conditions impairing DNA replication and genome integrity. Interestingly, a recent study provided evidence in favor of the activation of the Cu-import system in Saccharomyces cerevisiae in response to treatments with DNA-damaging agents (Dong et al., 2013). In this regard, our data supporting the KIN17-SPL7 interaction would reinforce the requirement of Cu in DNA metabolism and provide a first indication of how this coupling is achieved in plants. Furthermore, the characterization of the kin17-1 spl7-2 double mutant also shed some light on the biological relevance of the SPL7-KIN17 node to counteract oxidative stress and secure plant growth and development in response to Cu scarcity. Agarwal and colleagues (2006) described how Cu deficiency results in increased amounts of hydrogen peroxide and lipid peroxidation in wheat (Triticum aestivum). Several studies led to the conclusion that the photosynthetic electron transport chain (PETC) constitutes a link between Cu deficiency and oxidative stress. Ravet and Pilon (2013) suggested that a blockage in the PETC may provoke the photoreduction of oxygen at PSII and the incapacity to maintain balanced NADPH/NADP+ and reduced/oxidized glutathione pools. In fact, to fulfill the PC demand for Cu and maintain PETC functionality during Cu starvation, plants economize on the usage of Cu by replacing Cu/Zn SOD with an Fe isoform and simultaneously mitigating related reactive oxygen species (ROS) production (Abdel-Ghany et al., 2005). Therefore, limiting Cu to the chloroplast, by suppressing the chloroplastic Cu-transport P-ATPases PAA1 or PAA2 or the intracellular Cu-transport protein COPT5, provokes impairments in the stability of photosystems and decreases chlorophyll amounts (Shikanai et al., 2003; Abdel-Ghany et al., 2005; Garcia-Molina et al., 2011; Klaumann et al., 2011). But even though spl7-2 single mutants are supposed to suffer from inappropriate Cu uptake and reallocation as well as reduced antioxidant activities (e.g. inadequate substitution of the chloroplastic SODs; Yamasaki et al., 2009; Bernal et al., 2012), the phenotype did not show a dramatic alteration either in the anthocyanin content or in the lipid peroxidation rates. Both factors were clearly affected in the kin17-1 spl7-2 double mutant. Consequently, we propose an SPL7-overlapping and KIN17-dependent mechanism to alleviate the ROS damage and, thus, preservation of cell integrity and plant growth under Cu scarcity.
Abiotic stresses in plants predominantly affect reproductive growth, and for both Cu deficiency and oxidative stress, pollen development constitutes a particularly sensitive process (Dell, 1981; Wan et al., 2007; Higashitani, 2013). On the one hand, and consistent with a relative peak in SPL7 expression (AtGenExpress; Schmid et al., 2005), the plasma membrane-associated Cu-high affinity transport proteins (i.e. COPT1, COPT2, and COPT6) are particularly highly expressed in pollen (Honys and Twell, 2004; Sancenón et al., 2004; Jung et al., 2012; Garcia-Molina et al., 2013; Perea-García et al., 2013). Moreover, antisense-mediated knockdown of COPT1 in Arabidopsis causes the development of morphological aberrant pollen grains in a Cu-dependent manner (Sancenón et al., 2004). On the other hand, mutations that lead to impairments in the antioxidant barriers negatively impact the development of the male gametophyte and, eventually, pollen production. For instance, the anther-expressed MADS-domain homeotic factor, MADS3, regulates a cohort of genes related to ROS scavenging in rice (Oryza sativa; Hu et al., 2011). Thus, the male sterility in the mads3-4 mutant is associated with a reduced antioxidant response (Hu et al., 2011). Similarly, by mutation of the Arabidopsis CC-type glutaredoxins ROXY1 and ROXY2, their antioxidant activity was revealed to be essential in the adaxial lobes of anthers for the formation and differentiation of the tapetal and sporogenic tissues (Xing and Zachgo, 2008). In addition, a blockage in the biosynthesis of glutathione reduces pollen germination of the phytoalexin deficient2-1 mutant (Zechmann et al., 2011). Furthermore, studies in cotton (Gossypium hirsutum) plants uncovered that an increase in ROS constitutes the signal for the abortion of pollen associated with cytoplasmic male sterility (Jiang et al., 2007). Consequently, the semisterility displayed by the kin17-1 spl7-2 mutant due to severely compromised pollen production and viability under Cu scarcity reflects the importance of a coordinated KIN17-SPL7 action.
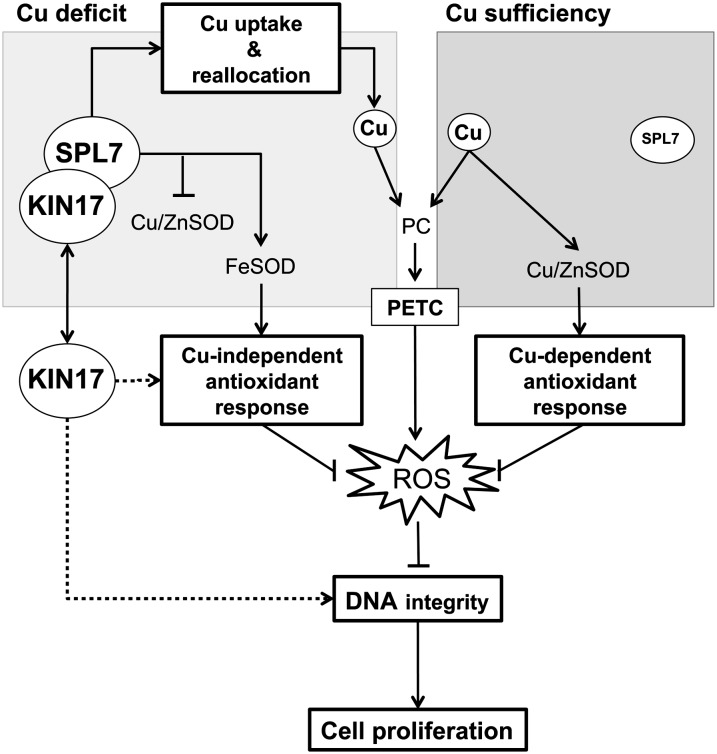
The data we present not only provide novel support for the importance of Cu in plant development but also evidence for a posttranslational mechanism to regulate SPL7 function during oxidative stress conditions. We propose a working model in which the Arabidopsis KIN17 protein physically interacts with the SPL7 transcription factor to reinforce SPL7 function during Cu starvation and to attenuate the consequences of oxidative stress (Fig. 10). Due to the pivotal role of photosynthesis, vegetative plant cells need to maintain a continuous pool of functional PC and antioxidant activities to cope with the ROS production related to PETC or adverse environmental conditions. PC demands can easily be fulfilled under Cu sufficiency, as well as Cu enzymes involved in the antioxidant response, most importantly the Cu/Zn SOD. However, given a hypothetic steady-state situation of Cu scarcity, a priority in Cu delivery to PC and other fundamental Cu proteins would limit the cellular Cu-dependent antioxidant capacity. In this scenario, the SPL7 transcription factor would stimulate the Cu-uptake system and promote a coordinated replacement of the chloroplastic Cu/Zn SOD by its Fe-dependent isoform. Consequently, the Fe SOD, in concert with other enzymes and compounds (e.g. glutaredoxins, glutathione, tocopherol, and ascorbic acid), would lead the Cu-independent antioxidant response. However, when rising above a certain threshold, ROS may cause DNA damage and, eventually, growth arrest. Hence, to maintain DNA integrity under such circumstances, and thus proper cell proliferation, we propose three different functional roles for KIN17. First, KIN17 would interact with SPL7 to reinforce the Cu starvation-induced reprogramming of the transcriptome to reinforce the cellular antioxidant system or additional yet-unknown Cu-dependent processes related to plant growth and development. Second, KIN17 would converge, direct or indirectly, with SPL7 in some antioxidant strategy aimed to mitigate oxidative stress. Third, albeit more speculatively, Arabidopsis KIN17 would contribute directly to the repair of DNA lesions, reflecting the conservation of a major function fulfilled by its orthologs in mammals (Fig. 10).
Figure 10.
A working model for the KIN17-SPL7 node function under Cu deficiency. ROS are constantly generated in plant cells, especially due to PETC activity. Increased levels of ROS cause DNA damage and eventually result in abrogation of cell proliferation. The availability of Cu determines which enzymatic antioxidant response plants activate to counteract the deleterious effect of ROS. Whereas Cu is used to fulfill Cu-dependent antioxidant responses led by the Cu/Zn SOD during Cu sufficiency, it must be economized for fundamentally important Cu proteins during Cu-deficient periods. Consequently, the SPL7 transcription factor promotes a Cu-uptake and -redistribution strategy to preferentially allocate Cu toward PC and thus maintain the PETC as functional. Additionally, SPL7 also coordinates the Cu/Zn SOD substitution by the Fe SOD isoform to ensure a Cu-independent antioxidant response. In the case of high ROS levels, we propose that Arabidopsis KIN17 acts at three levels to counteract this scenario: (1) KIN17 would associate with SPL7 either to enhance the Cu-starvation response and reinforce the cellular antioxidant system or to protect additional yet-unknown Cu-dependent processes related to plant growth and development; (2) KIN17 would converge, directly or indirectly, with SPL7 to maintain some Cu-independent antioxidant system; and (3) KIN17 would also be directly involved in repairing DNA lesions, as reported in mammals.
MATERIALS AND METHODS
Plant Growth and Manipulation
For all the experiments, Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used as the wild type. Seeds were stratified at 4°C for 2 d prior to being sown on soil and regularly watered or treated with a 5 mm CuSO4 solution. For in vitro culture, seeds were surface sterilized with sequential washes in 70% (v/v) ethanol (5 min), bleach (5 min), water (twice for 2 min each), resuspended in 0.1% (w/v) agar, and sown on plates containing 1/2 MS (Sigma-Aldrich) supplemented with 1% (w/v) Suc or as indicated. Hydroponic cultures were performed with seedlings germinated in Eppendorf tubes filled with 0.1× Hoagland solution with 0.7% (w/v) agar that were transferred to black boxes containing modified 0.1× Hoagland solution as described by Hermans and Verbruggen (2005) without Cu or with 1 µm CuSO4. In all cases, long-day conditions (16 h of light, 20°C–23°C/8 h of darkness, 16°C) were applied.
The kin17-1 line corresponds to a T-DNA insertion mutant in the Columbia background (N677041 from the Nottingham Arabidopsis Stock Centre). The spl7-2 mutant has been described previously (Bernal et al., 2012). To generate the double mutant kin17-1 spl7-2, pollen from kin17-1 was manually applied to spl7-2 emasculated flowers.
Stable Arabidopsis transgenic lines were generated by means of the Agrobacterium tumefaciens GV3101 (pMP90RK) strain using floral dipping (Koncz and Schell, 1986; Clough and Bent, 1998). For transient transformation assays, young tobacco (Nicotiana benthamiana) plants grown on soil under long-day conditions were used. Briefly, nonsaturated cultures of A. tumefaciens harboring the desired constructs or the silencing-suppressor p19 plasmid (Voinnet et al., 2003) were pelleted, diluted to an optical density at 600 nm of 0.2 in infiltration buffer (0.5% [w/v] d-Glc, 10 mm MES, 10 mm MgCl2, and 0.1 mm acetosyringone), and coinfiltrated in young tobacco leaves.
Y2H Screen
The Y2H assay was performed by Hybrigenics Services using a fragment of SPL7 (amino acid residues 133–762) as bait to screen a random-primed complementary DNA (cDNA) prey library prepared from 1-week-old Arabidopsis seedlings.
Constructs
The Gateway recombination cloning technology was used to generate all recombinant vector constructs described in this work. First, PCR products were cloned into the pDONR207 entry vector with GATEWAY BP CLONASE II enzyme mix (Invitrogen), and the subsequent products were recombined into a suitable destination vector by means of GATEWAY LR CLONASE II enzyme mix (Invitrogen). All oligonucleotides used for PCR are listed in Supplemental Table S1.
The entire SPL7 and KIN17 coding sequences were amplified with the specific oligonucleotides SPL7-ATG-F/SPL7-TGA-R and KIN17-ATG-F/KIN17-TGA-R, respectively, and BP recombined in pDONR207 to generate pDONR207::SPL7 and pDONR207::KIN17. Next, to generate N-terminal fusions to GFP (GFP::SPL7 and GFP::KIN17), both vectors were LR recombined into the pMDC43 vector (Curtis and Grossniklaus, 2003). pDONR207::KIN17 was also LR recombined into pALLIGATOR2 (Bensmihen et al., 2004) to fuse KIN17 to the human influenza HA epitope tag (HA::KIN17). For BiFC assays, the pDONR207::SPL7 and pDONR207::KIN17 entry vectors were recombined into the pYFN43 and pYFC43 destiny vectors (Belda-Palazón et al., 2012), respectively, to generate an N-terminal translational fusion of the two halves of YFP to SPL7 and KIN17 (nYFP::SPL7 and cYFP::KIN17).
The KIN17 promoter was amplified with the specific primer pair pKIN17-F/pKIN17-R and inserted in pDONR207 to generate pDONR207::pKIN17. Subsequently, it was LR recombined into pGWFS7 to insert it in front of GFP and the uidA gene encoding the GUS (Karimi et al., 2002). Similarly, a DNA fragment containing pKIN17::KIN17 was amplified with the primer pair pKIN17-F/KIN17-TGA-R, introduced into pDONR207, and finally in pMDC123 (Curtis and Grossniklaus, 2003) to complement the kin17-1 mutant.
Coimmunoprecipitation
For coimmunoprecipitation assays, tobacco leaves were transiently transformed with the pALLIGATOR2::KIN17 and pMDC43::SPL7 constructs. Total proteins from 3-d-after-infiltration leaves were extracted and limited cross linked by grinding frozen material in coimmunoprecipitation buffer (10 mm PIPES-KOH, pH 7, 50 mm NaCl, 0.5 mm EDTA, pH 8, 0.5% [v/v] Triton X-100, 1% [v/v] formaldehyde, and 1× Complete Protease Inhibitor Cocktail [Roche Diagnostics]). Samples were incubated on ice for 5 min and rapidly centrifuged at maximum speed at 4°C in a bench centrifuge for 5 min. The cross linking was stopped by adding 0.4 volumes of 2 m Gly to the supernatants. A total of 1.5 µg of anti-HA high affinity (3F10; Roche Diagnostics) was added to 1 mL of protein extract and rotated at 4°C for 2 h. Then, 50 µL of Protein G Mag Sepharose Xtra (GE Healthcare) equilibrated with coimmunoprecipitation buffer without Triton X-100, formaldehyde, and protease inhibitor was added to extracts and rotated overnight at 4°C. Beads were washed three times with 1 mL of coimmunoprecipitation buffer supplemented with 300 mm NaCl, 0.1% (v/v) Triton X-100, and 1× Complete Protease Inhibitor Cocktail (Roche Diagnostics) and boiled at 95°C for 10 min with 50 µL of 4× SDS-PAGE loading buffer. The obtained supernatants were loaded into a 10% (w/v) SDS-PAGE device, blotted onto a polyvinylidene difluoride membrane, probed with anti-GFP-horseradish peroxidase (HRP; 1:1,000; Molecular Probes, Life Technologies Europe) or anti-HA-HRP (1:1,000; Roche Diagnostics) antibodies, and developed with the Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare).
Subcellular Localization and BiFC Assay
To determine the subcellular localization of KIN17 and SPL7, the pMDC43::KIN17 and pMDC43::SPL7 vectors were transiently expressed in tobacco leaves, and 2 d after infiltration, small pieces were excised and visualized on an LSM 700 confocal laser microscope (Carl Zeiss) using specific filter settings to detect the GFP signal. For BiFC assays, the pYFN43::SPL7 and pYFC43::KIN17 destiny vectors were individually expressed or coexpressed in tobacco leaves, and 2-d-transformed leaves were observed using confocal imaging to assess the restoration of the YFP signal.
Gene Expression Analysis by Quantitative Real-Time PCR
For gene expression assays, total RNA from 7-d-old seedlings or selected organs was prepared with the Spectrum Plant Total RNA Kit (Sigma-Aldrich) according to the manufacturer’s recommendations. The RNA quality and integrity were evaluated by capillary electrophoresis in an Agilent 2100 Bioanalyzer (Agilent Technologies). In all cases, the RNA integrity number was above 8. RNA was treated with DNase I recombinant RNase-free (Roche Diagnostics) and reverse transcribed to cDNA with the SuperScript II reverse transcriptase (Invitrogen). qPCR analysis was carried out in an iQ5 Real-Time PCR Detection System (Bio-Rad Laboratories) with EvaGreen (Biotium) and specific primers (Supplemental Table S2) using an initial cycle at 95°C for 3 min and 40 cycles consisting of 95°C for 10 s, 58°C for 20 s, and 72°C for 20 s.
Pollen Analysis
To count pollen, 25 to 30 stamens from flowers in position 5 to 10 were manually dissected and disrupted using a Teflon pestle in an end volume of 400 µL of phosphate-buffered saline and 20% (v/v) glycerol. Pollen was pelleted by centrifugation at maximum speed for 10 min and then resuspended with 100 µL of phosphate-buffered saline, 20% (v/v) glycerol, and lacto-phenol-fuchsine (40% [v/v] glycerol, 20% [v/v] lactic acid, 20% [v/v] phenol, and 0.1% [w/v] acid fuchsine). For viability assays, pollen of five to 10 freshly opened flowers was collected on 8% (w/v) Suc and 0.001% (w/v) FDA, vortexed, recovered by centrifuging at maximum speed for 5 min, and resuspended in 50 µL of the Suc-FDA solution. Finally, 5 µL of the pollen suspensions was spotted on a 1% (w/v) agar plate and observed with a binocular microscope for UV-excited fluorescence.
Miscellaneous Methods
Histochemical GUS assay was performed in pKIN17::GUS plants as described by Jefferson et al. (1987). Genomic DNA was extracted from leaflets using extraction buffer (200 mm Tris-HCl, pH 7.5, 250 mm NaCl, 25 mm EDTA, and 0.5% [w/v] SDS) and subsequently with isopropanol. T-DNA mutants were genotyped using the specific oligonucleotides indicated in Supplemental Table S3. Anthocyanin content in leaves was quantified according to Mita et al. (1997). Briefly, pigments from 20 mg of frozen tissue were extracted in 1 mL of methanol and 1% (v/v) HCl and incubated at 4°C for 6 h, and the anthocyanin content was spectrophotometrically ascertained from the supernatants by the formula A530 – 1/4 A657 and normalized to the fresh weight. To infer the lipid peroxidation, frozen rosette leaves were ground on malondialdehyde extraction buffer (15% [v/v] TCA, 0.37% [w/v] 2-thiobarbituric acid, 0.24 n HCl, and 0.001% [w/v] butylated hydroxytoluene) and boiled at 80°C for 1 h to generate malondialdehyde derivatives that were determined by the formula (A535 − A600)/(ε 1.56 × 105 M−1 cm−1) and normalized to fresh weight. Multiple alignments were carried out with the MacVector software package (Accelrys). Statistical analysis of data was performed using Excel (Microsoft). Student’s t test was used to determine statistically significant differences with P < 0.05 as the level of significance.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. SPL7 interacts with KIN17 in a yeast two-hybrid assay.
Supplemental Figure S2. Multiple sequence alignment of different KIN17 homologs.
Supplemental Figure S3. Bimolecular fluorescence complementation assay with SPL7 and KIN17.
Supplemental Figure S4. Isolation and characterisation of the double mutant kin17-1 spl7-2.
Supplemental Table S1. Oligonucleotides used for cloning.
Supplemental Table S2. Oligonucleotides used for quantitative real-time PCR.
Supplemental Table S3. Oligonucleotides used to genotype T-DNA mutants.
Acknowledgments
We thank Elmon Schmelzer (Central Microscopy service unit at the Max Planck Institute for Plant Breeding Research) for his advice and instructions on confocal microscopy and Arne Grande, Susanne Höhmann, and Rita Berndtgen (Max Planck Institute for Plant Breeding Research) for their technical support in generating SPL7 constructs and qPCR, and François Parcy (Commissariat à l’Energie Atomique-Centre National de la Recherche Scientifique) and Alejandro Ferrando (Instituto de Biología Molecular y Celular de Plantas-Consejo Superior de Investigaciones Científicas) for kindly providing pALLIGATOR2 and pYFN43 vectors, respectively.
Glossary
- Cu
copper
- PC
plastocyanin
- Y2H
yeast two-hybrid
- HA
hemagglutinin
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- 1/2 MS
one-half-strength Murashige and Skoog
- qPCR
quantitative PCR
- T-DNA
transfer DNA
- BCS
bathocuproine disulfonate
- –Cu
very mild copper deficiency
- +Cu
extra copper
- FDA
fluorescein diacetate
- PETC
photosynthetic electron transport chain
- ROS
reactive oxygen species
- Zn
zinc
- Fe
iron
- SOD
superoxide dismutase
- cDNA
complementary DNA
- HRP
horseradish peroxidase
References
- Abdel-Ghany SE, Müller-Moulé P, Niyogi KK, Pilon M, Shikanai T. (2005) Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17: 1233–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Ghany SE, Pilon M. (2008) MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J Biol Chem 283: 15932–15945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S, Sairam RK, Meena RC, Tyagi A, Srivastava GC. (2006) Effect of excess and deficient levels of iron and copper on oxidative stress and antioxidant enzymes activity in wheat. J Plant Sci 1: 86–97 [Google Scholar]
- Angulo JF, Moreau PL, Maunoury R, Laporte J, Hill AM, Bertolotti R, Devoret R. (1989) KIN, a mammalian nuclear protein immunologically related to E. coli RecA protein. Mutat Res 217: 123–134 [DOI] [PubMed] [Google Scholar]
- Belda-Palazón B, Ruiz L, Martí E, Tárraga S, Tiburcio AF, Culiáñez F, Farràs R, Carrasco P, Ferrando A. (2012) Aminopropyltransferases involved in polyamine biosynthesis localize preferentially in the nucleus of plant cells. PLoS ONE 7: e46907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensmihen S, To A, Lambert G, Kroj T, Giraudat J, Parcy F. (2004) Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett 561: 127–131 [DOI] [PubMed] [Google Scholar]
- Bernal M, Casero D, Singh V, Wilson GT, Grande A, Yang H, Dodani SC, Pellegrini M, Huijser P, Connolly EL, et al. (2012) Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. Plant Cell 24: 738–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biard DS, Miccoli L, Despras E, Frobert Y, Creminon C, Angulo JF. (2002) Ionizing radiation triggers chromatin-bound kin17 complex formation in human cells. J Biol Chem 277: 19156–19165 [DOI] [PubMed] [Google Scholar]
- Biard DS, Miccoli L, Despras E, Harper F, Pichard E, Créminon C, Angulo JF. (2003) Participation of kin17 protein in replication factories and in other DNA transactions mediated by high molecular weight nuclear complexes. Mol Cancer Res 1: 519–531 [PubMed] [Google Scholar]
- Birkenbihl RP, Jach G, Saedler H, Huijser P. (2005) Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J Mol Biol 352: 585–596 [DOI] [PubMed] [Google Scholar]
- Burkhead JL, Reynolds KA, Abdel-Ghany SE, Cohu CM, Pilon M. (2009) Copper homeostasis. New Phytol 182: 799–816 [DOI] [PubMed] [Google Scholar]
- Carlier L, Couprie J, le Maire A, Guilhaudis L, Milazzo-Segalas I, Courçon M, Moutiez M, Gondry M, Davoust D, Gilquin B, et al. (2007) Solution structure of the region 51-160 of human KIN17 reveals an atypical winged helix domain. Protein Sci 16: 2750–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell B. (1981) Male sterility and anther wall structure in copper-deficient plants. Ann Bot (Lond) 48: 599–608 [Google Scholar]
- Dong K, Addinall SG, Lydall D, Rutherford JC. (2013) The yeast copper response is regulated by DNA damage. Mol Cell Biol 33: 4041–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Molina A, Andrés-Colás N, Perea-García A, Del Valle-Tascón S, Peñarrubia L, Puig S. (2011) The intracellular Arabidopsis COPT5 transport protein is required for photosynthetic electron transport under severe copper deficiency. Plant J 65: 848–860 [DOI] [PubMed] [Google Scholar]
- Garcia-Molina A, Andrés-Colás N, Perea-García A, Neumann U, Dodani SC, Huijser P, Peñarrubia L, Puig S. (2013) The Arabidopsis COPT6 transport protein functions in copper distribution under copper-deficient conditions. Plant Cell Physiol 54: 1378–1390 [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JM. (1984) Lipid peroxidation, oxygen radicals, cell damage, and antioxidant therapy. Lancet 1: 1396–1397 [DOI] [PubMed] [Google Scholar]
- Hermans C, Verbruggen N. (2005) Physiological characterization of Mg deficiency in Arabidopsis thaliana. J Exp Bot 56: 2153–2161 [DOI] [PubMed] [Google Scholar]
- Higashitani A. (2013) High temperature injury and auxin biosynthesis in microsporogenesis. Front Plant Sci 4: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himelblau E, Mira H, Lin SJ, Culotta VC, Peñarrubia L, Amasino RM. (1998) Identification of a functional homolog of the yeast copper homeostasis gene ATX1 from Arabidopsis. Plant Physiol 117: 1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D. (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Liang W, Yin C, Cui X, Zong J, Wang X, Hu J, Zhang D. (2011) Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23: 515–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Zhang X, Zhu Y, Zhu W, Xie H, Wang X. (2007) Metabolism of reactive oxygen species in cotton cytoplasmic male sterility and its restoration. Plant Cell Rep 26: 1627–1634 [DOI] [PubMed] [Google Scholar]
- Jung HI, Gayomba SR, Rutzke MA, Craft E, Kochian LV, Vatamaniuk OK. (2012) COPT6 is a plasma membrane transporter that functions in copper homeostasis in Arabidopsis and is a novel target of SQUAMOSA promoter-binding protein-like 7. J Biol Chem 287: 33252–33267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche P, Pinon-Lataillade G, Mauffrey P, Faucher C, Biard DS, Angulo JF. (1997) Overexpression of kin17 protein forms intranuclear foci in mammalian cells. Biochimie 79: 599–606 [DOI] [PubMed] [Google Scholar]
- Kannouche P, Pinon-Lataillade G, Tissier A, Chevalier-Lagente O, Sarasin A, Mezzina M, Angulo JF. (1998) The nuclear concentration of kin17, a mouse protein that binds to curved DNA, increases during cell proliferation and after UV irradiation. Carcinogenesis 19: 781–789 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Klaumann S, Nickolaus SD, Fürst SH, Starck S, Schneider S, Ekkehard Neuhaus H, Trentmann O. (2011) The tonoplast copper transporter COPT5 acts as an exporter and is required for interorgan allocation of copper in Arabidopsis thaliana. New Phytol 192: 393–404 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genetics 204: 383–396 [Google Scholar]
- Kropat J, Tottey S, Birkenbihl RP, Depège N, Huijser P, Merchant S. (2005) A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc Natl Acad Sci USA 102: 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuper J, Llamas A, Hecht HJ, Mendel RR, Schwarz G. (2004) Structure of the molybdopterin-bound Cnx1G domain links molybdenum and copper metabolism. Nature 430: 803–806 [DOI] [PubMed] [Google Scholar]
- Kyrpides NC, Woese CR, Ouzounis CA. (1996) KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem Sci 21: 425–426 [DOI] [PubMed] [Google Scholar]
- le Maire A, Schiltz M, Stura EA, Pinon-Lataillade G, Couprie J, Moutiez M, Gondry M, Angulo JF, Zinn-Justin S. (2006) A tandem of SH3-like domains participates in RNA binding in KIN17, a human protein activated in response to genotoxics. J Mol Biol 364: 764–776 [DOI] [PubMed] [Google Scholar]
- Masson C, Menaa F, Pinon-Lataillade G, Frobert Y, Chevillard S, Radicella JP, Sarasin A, Angulo JF. (2003) Global genome repair is required to activate KIN17, a UVC-responsive gene involved in DNA replication. Proc Natl Acad Sci USA 100: 616–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin A, Timchenko T, Ménissier-de Murcia J, Schreiber V, Angulo JF, de Murcia G, Devoret R. (1994) Kin17, a mouse nuclear zinc finger protein that binds preferentially to curved DNA. Nucleic Acids Res 22: 4335–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miccoli L, Frouin I, Novac O, Di Paola D, Harper F, Zannis-Hadjopoulos M, Maga G, Biard DS, Angulo JF. (2005) The human stress-activated protein kin17 belongs to the multiprotein DNA replication complex and associates in vivo with mammalian replication origins. Mol Cell Biol 25: 3814–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira H, Martínez-García F, Peñarrubia L. (2001) Evidence for the plant-specific intercellular transport of the Arabidopsis copper chaperone CCH. Plant J 25: 521–528 [DOI] [PubMed] [Google Scholar]
- Mita S, Murano N, Akaike M, Nakamura K. (1997) Mutants of Arabidopsis thaliana with pleiotropic effects on the expression of the gene for beta-amylase and on the accumulation of anthocyanin that are inducible by sugars. Plant J 11: 841–851 [DOI] [PubMed] [Google Scholar]
- Ohyama T (2005) Curved DNA and transcription in eukaryotes. In T Ohyama, ed, DNA Conformation and Transcription. Springer, New York, pp 66–74 [Google Scholar]
- Perea-García A, Garcia-Molina A, Andrés-Colás N, Vera-Sirera F, Pérez-Amador MA, Puig S, Peñarrubia L. (2013) Arabidopsis copper transport protein COPT2 participates in the cross talk between iron deficiency responses and low-phosphate signaling. Plant Physiol 162: 180–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig S, Andrés-Colás N, García-Molina A, Peñarrubia L. (2007) Copper and iron homeostasis in Arabidopsis: responses to metal deficiencies, interactions and biotechnological applications. Plant Cell Environ 30: 271–290 [DOI] [PubMed] [Google Scholar]
- Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O’Halloran TV. (1999) Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science 284: 805–808 [DOI] [PubMed] [Google Scholar]
- Ravet K, Pilon M. (2013) Copper and iron homeostasis in plants: the challenges of oxidative stress. Antioxid Redox Signal 19: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB. (1999) A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science 283: 996–998 [DOI] [PubMed] [Google Scholar]
- Sancenón V, Puig S, Mateu-Andrés I, Dorcey E, Thiele DJ, Peñarrubia L. (2004) The Arabidopsis copper transporter COPT1 functions in root elongation and pollen development. J Biol Chem 279: 15348–15355 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Shikanai T, Müller-Moulé P, Munekage Y, Niyogi KK, Pilon M. (2003) PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell 15: 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, Kropat J, Malasarn D, Grossoehme NE, Chen X, Giedroc DP, Merchant SS. (2010) The CRR1 nutritional copper sensor in Chlamydomonas contains two distinct metal-responsive domains. Plant Cell 22: 4098–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wan C, Li S, Wen L, Kong J, Wang K, Zhu Y. (2007) Damage of oxidative stress on mitochondria during microspores development in Honglian CMS line of rice. Plant Cell Rep 26: 373–382 [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang D, Chu JY, Boyle P, Wang Y, Brindle ID, De Luca V, Després C. (2012) The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep 1: 639–647 [DOI] [PubMed] [Google Scholar]
- Xing S, Zachgo S. (2008) ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J 53: 790–801 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T. (2009) SQUAMOSA Promoter Binding Protein-Like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21: 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Inoue M, Watanabe S, Tateno M, Seki M, Shinozaki K, Yokoyama S. (2008) Structures and evolutionary origins of plant-specific transcription factor DNA-binding domains. Plant Physiol Biochem 46: 394–401 [DOI] [PubMed] [Google Scholar]
- Zechmann B, Koffler BE, Russell SD. (2011) Glutathione synthesis is essential for pollen germination in vitro. BMC Plant Biol 11: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]