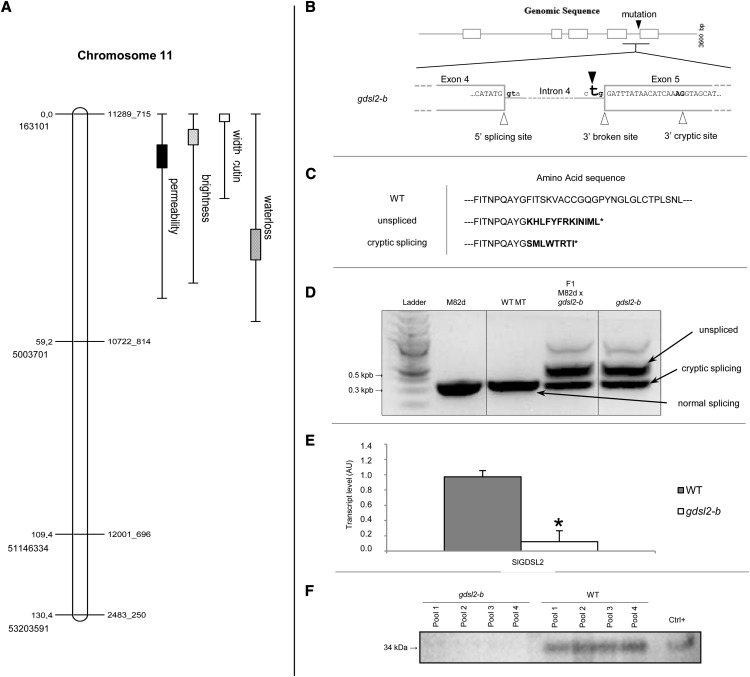
Figure 7.
Mapping of the P15C12 glossy mutant and characterization of the mutation. A, The four cuticle-related traits (brightness, water loss, permeability, cutin width) were mapped to chromosome 11 between SNP markers 11289_715 and 10722_814. Genetic distance (in centimorgans) and physical position (ITAG2.40) are indicated on the left. Markers are indicated on the right. 1-LOD and 2-LOD support intervals of each QTL are marked by boxes and thin bars, respectively. B, SlGDSL2 gene splicing model. Exons, introns, and splicing sites (in bold) are represented by white boxes, gray lines, and white triangles, respectively. The black triangle shows the A to T mutation in the 3′ consensus splice site of intron 4, which results in a broken splicing site in the gdsl2-b mutant. The 3′ cryptic splicing site is shown on the right. C, Amino acid sequences deduced from native SlGDSL2 transcript (wild-type plant, SlGDSL2 gene), unspliced transcript (in-frame reading through intron 4 in the gdsl2-b mutant), and alternative cryptic site transcript (splicing of intron 4 at 3′ cryptic site in exon 5 in the gdsl2-b mutant). Incorrect amino acids are indicated in bold. D, Electrophoretic analysis of the SlGDSL2 transcripts in M82dwarf (M82d), wild-type ‘Micro-Tom’ (WT MT), gdsl2-b mutant, and F1 hybrid between M82d and gdsl2-b. E, Real-time reverse-transcription PCR analysis of SlGDSL2 expression in 20 DPA fruits in wild-type ‘Micro-Tom’ (gray box) and gdsl2-b mutant (white box). Mean values of three biological replicates are shown with sd. Asterisk indicates significant difference from wild-type fruits (t test, P < 0.01). F, Immunoblot analysis of GDSL2 protein in four independent pools of gdsl2-b and wild-type epidermis. Positive control is a recombinant GDSL extract (Girard et al., 2012). AU, Arbitrary units; Ctrl, control; LOD, log of the odds; QTL, quantitative trait loci.