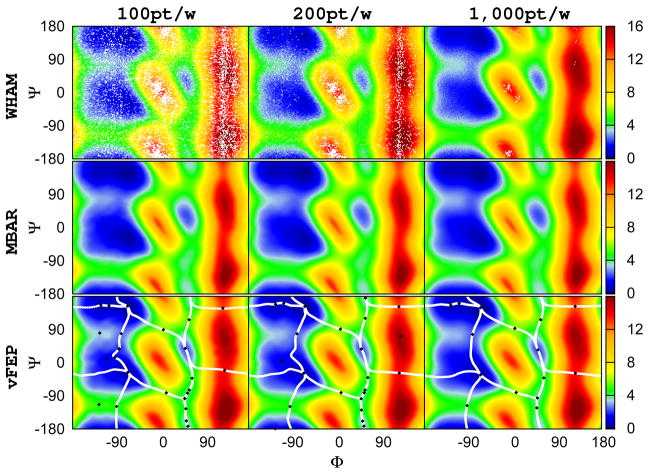
Figure 3.
The 2D (φ, ψ) free energy profile of alanine dipeptide calculated from different methods using 24×24 (576) windows: WHAM, MBAR/gKDE, and vFEP results are shown in the upper, middle, and bottom rows respectively. Results obtained from different 100, 200, and 1000 points per window are shown in the left, middle, and right columns respectively. White space in the WHAM results indicate empty histogram bins. MBAR/gKDE is able to deliver good results while WHAM still cannot converge in all regions, even with 1000 points per window. The 2D-vFEP results with 200 and 1000 points are essentially the same, and the results with 100 points per window extremely similar in qualitative terms. All angles are in degrees. The color scheme of the free energy profile is shown at the right side color bar with units in kcal/mol. The black diamonds and the white lines shown in the vFEP contour maps are the saddle/minimum points and the possible paths found by the NEB method, respectively.