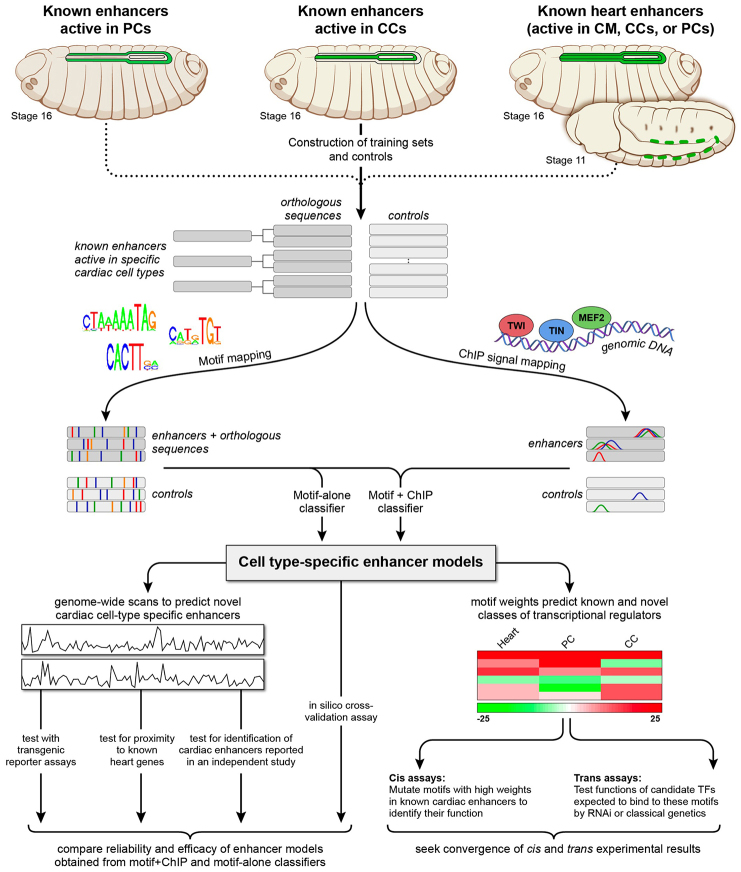
Fig. 1.
Schematic overview of the computational and experimental strategy utilized in this study. Training sets of enhancers expressed in different cardiac cell types (increased in size through phylogenetic profiling) and control sequences were compiled and subsequently scanned to map sequence features corresponding to known binding site motifs collected from public databases and to in vivo TF binding signals obtained from published ChIP data profiles. Classifiers were built to create enhancer models that discriminate cell type-specific enhancers from respective controls. For each cell type, two classifiers were independently constructed: one based solely on motif features (‘motif-alone’) and the other on motif features and ChIP signals (‘motif+ChIP′). The enhancer models were used to scan the Drosophila genome to identify novel cell-specific enhancers similar to the training sets, and the reliability and efficacy of the motif-alone and motif+ChIP classifiers for different cardiac cell types were examined and compared. Sequence features positively associated with computational classification were examined further using cis and trans in vivo experimental assays to identify and determine the functional roles of both the binding motifs and their associated regulatory TFs.