Figure 7.
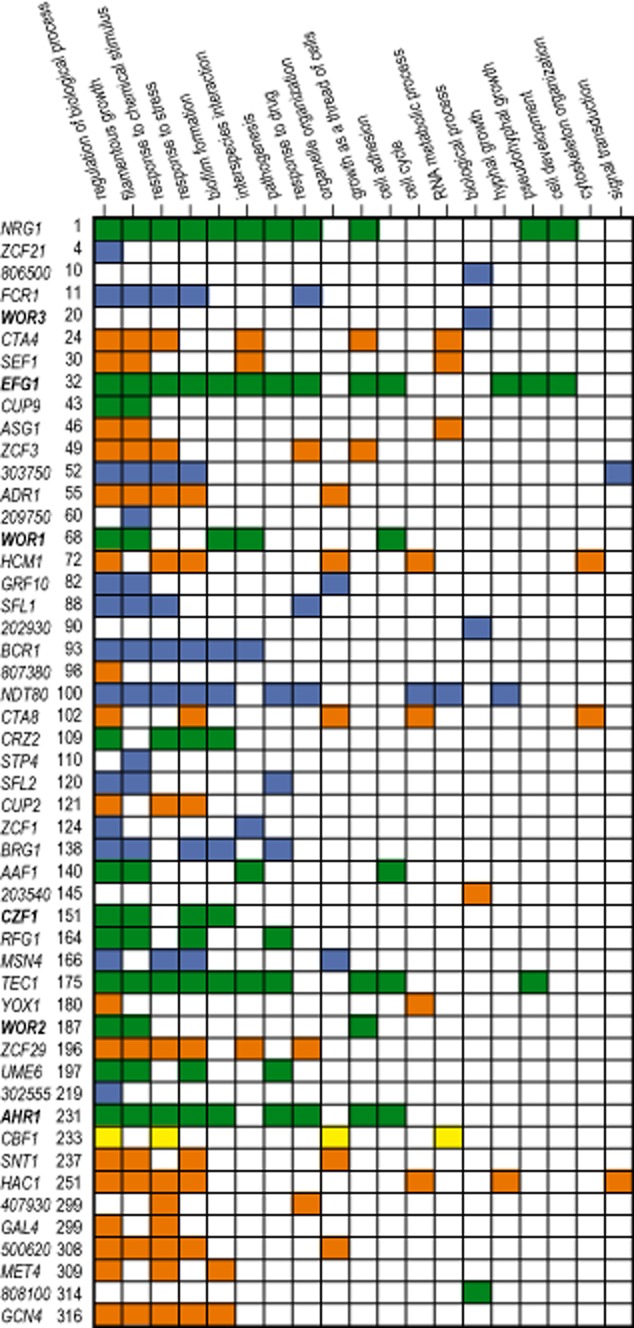
Similarities and differences between targets of Efg1 in C. parapsilosis and C. albicans. The top 50 transcription factors bound by Efg1 in C. parapsilosis are listed. The names of the C. albicans orthologues were used where possible; six-digit gene names (missing the suffix ‘CPAR2’) are used where only a C. albicans generic identifier is available (i.e. orf19). The gene name is followed by the rank from the ChIP-seq analysis (Table S2A). Processes associated with the 50 genes were determined using GO Slim Mapper at the Candida Genome Database (Inglis et al., 2012). Efg1 targets conserved in C. albicans are shown in green [identified by both Lassak et al. (2011) and Nobile et al. (2012)], yellow [identified by Lassak et al. (2011) only] and blue [identified by Nobile et al. (2012) only]. Orange boxes indicate that the relevant transcription factor is bound by Efg1 in C. parapsilosis only. Only processes including at least two genes were included. Genes highlighted in bold are part of the circuit regulating the white/opaque switch in C. albicans.
