Figure 1.
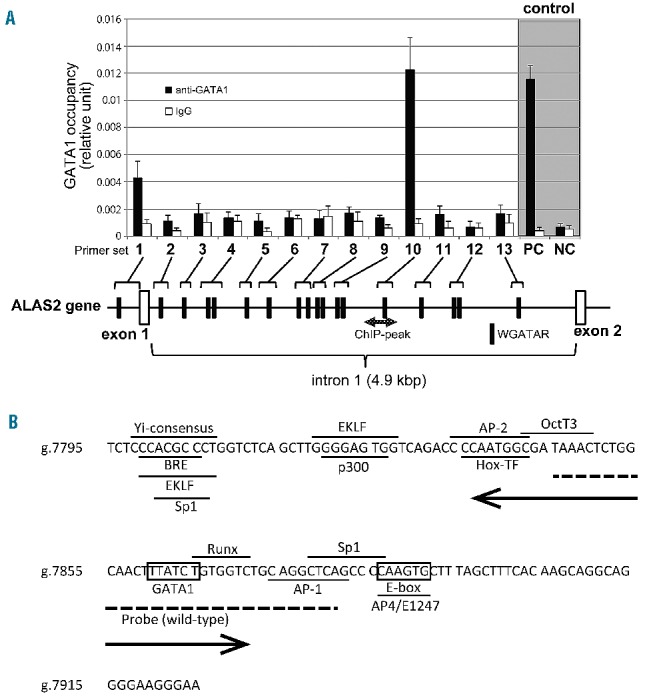
Identification of a functional GATA1 element in the first intron of the ALAS2 gene. (A) Chromatin immunoprecipitation assay. Fragmented genomic DNA segments were immunoprecipitated with anti- GATA1 antibody or control IgG, and then precipitated fragments were quantified using real-time PCR as described in the Online Supplementary Methods. PC or NC indicates positive control or negative control, respectively, for the ChIP assay using anti-GATA1 in K562 cells.22 One GATA element is present in the proximal promoter region and 17 GATA elements in the first intron (black symbols). The shaded double arrow indicates the region corresponding to ChIP-peak. (B) Nucleotide sequence of ChIPmini. The GATA binding site, ALAS2int1GATA, is located in the center of ChIPmini (boxed). A box also indicates the consensus for E-box that is bound by Scl/TAL1.22 The sequence of ChIPmini was further analyzed for putative transcription factor binding sites using GeneQuest software (DNASTAR Inc., Madison, WI, USA), and the results are indicated by the horizontal bar. Yi-consensus, Yi transcription factor consensus site;33 BRE, transcription factor IIB binding site;34 EKLF, erythroid/Kruppel-like factor consensus site;35 Sp1, stimulatory protein 1 binding site;36 P300, P300 transcriptional coactivator consensus site;37 AP-2, AP-2 beta consensus site;38 Hox-TF, C1 element binding factor binding site;39 OctT3, OctT3 binding site;40 Runx, Runx proteins binding site;41 AP-1, activator protein 1 binding site;42 and AP4/E1247, AP4/E1247 binding site.43 The sequence for the wild-type probe used in the EMSA is indicated by a dashed line. A double arrow indicates the deleted region of the delGATA mutation.
