Figure 6.
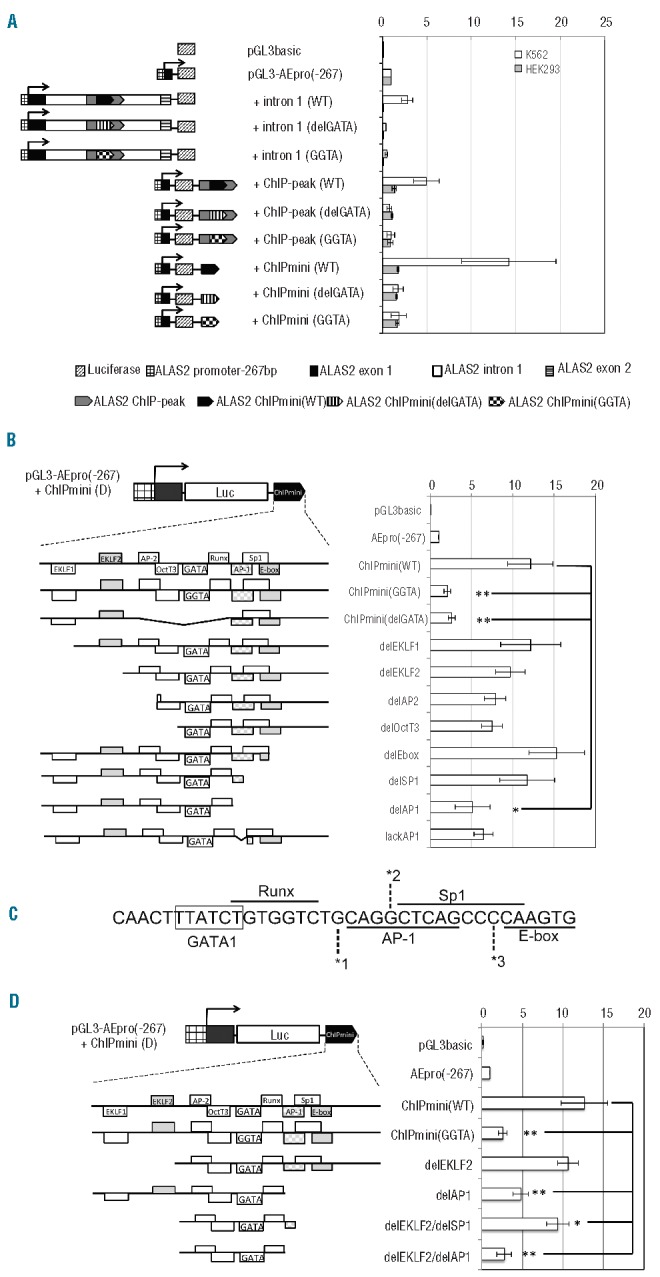
Identification of cis-elements essential for the erythroid-specific enhancer activity of ChIPmini. (A) Effect of each mutation of ALAS2int1GATA on the enhancer activity of ALAS2 ChIPmini. The region corresponding to +intron1, ChIP-peak or ChIPmini, derived from proband 1 or proband 3, was subcloned into pGL3-AEpro(−267) to construct the reporter vector containing the GGTA mutation or the deletion of ALAS2int1GATA, respectively. (B) Effect of the deletion at the 5′- or 3′-flanking region of ALAS2int1GATA on the enhancer activity of ChIPmini. The 5′- and 3′-flanking regions of ALAS2int1GATA contain potential transcription factor-binding sites (cis-elements), and a portion of each flanking region was deleted, as schematically shown. The enhancer activity of each deletion mutant was determined in K562 erythroleukemia cells. (C) The nucleotide sequence of the 3′-flanking region of ALAS2int1GATA. Note that the Sp1 site overlaps the AP-1 site and E-box. Each number, *1, *2 or *3, indicates the nucleotide at the 3′ end of the deletion mutant, delAP1, delSP1 or delE-box, respectively. Thus, delSP1 also lacks the 3′ portion of the AP-1 site. (D) Effect of deletion of the 5′- and 3′-flanking regions of ALAS2int1GATA on the enhancer activity of ChIPmini. The construct, delEKLF2/delSP1, lacks two EKLF sites in the 5′-flanking region and both the Sp1 element and E-box in the 3′-flanking region. The AP-1 element at the 3′-flanking region was deleted from delEKLF2/delSP1, yielding delEKLF2/delAP1. Results are expressed as relative activity compared to that of pGL3-AEpro(−267), and are presented as the mean ± SD of at least three independent experiments.
