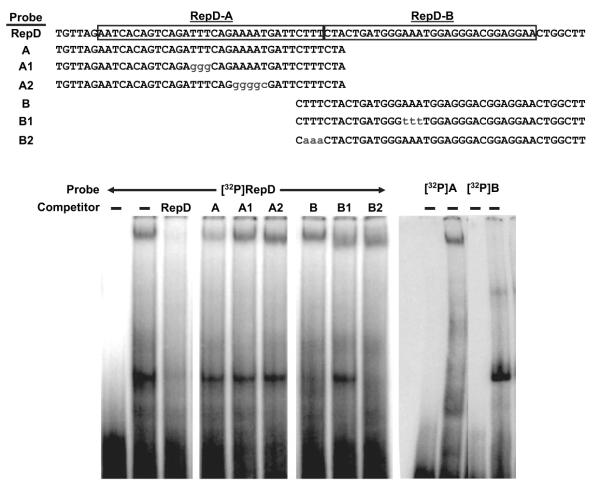
Figure 5.
Protein factors in endothelial cell nuclei bind RepD sequences. Nuclear proteins were extracted from porcine aortic endothelial cells and incubated with 32P-labeled oligonucleotide probes derived from the RepD sequence as shown (top). A control reaction contained labeled probe without protein (first lane). The protein–DNA complexes were resolved by nondenaturing polyacrylamide gel electrophoresis and detected on autoradiography film. The experiment revealed that 2 protein–DNA complexes form simultaneously on the radio-labeled RepD sequence. The slower-migrating complex is diminished when cold competitor oligonucleotide probes from intact RepD or from RepD-A are included but not when cold RepD-A sequences include the specific mutations shown. The faster-migrating complex is diminished by cold RepD or RepD-B oligonucleotides but not when RepD-B contains the mutation shown. Protein–DNA complexes formed with 32P-labeled oligonucleotide probes derived from RepD-A or RepD-B confirm that the slower-migrating complex forms specifically with the RepD-A sequence and that the faster-migrating complex forms specifically with the RepD-B sequence (right). Data are representative of 3 experiments.