Fig. 2.
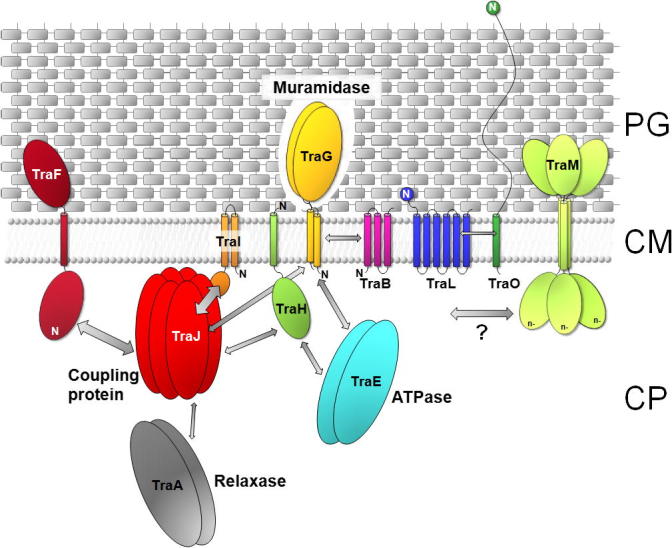
Model of localization and interactions of the pIP501 encoded T4SS proteins. Decreased shading of PG symbolizes TraG-mediated local opening of PG. The localization and orientation of the T4SS proteins is based on in silico predictions and localization studies (Arends et al., 2013; Goessweiner-Mohr et al., 2013). The N-terminus of the transfer proteins is marked (N). Arrows indicate protein–protein interactions determined by yeast-two-hybrid studies and validated by pull-down assays (Abajy et al., 2007), as well as interactions found by using the thermofluor method (Goessweiner-Mohr, N. et al., unpublished data). The VirB8-like T4S protein TraM is marked with a question mark as no protein–protein interactions have been detected so far. The thickness of the arrows marks the strength of the detected interactions. The putative function of key members of the pIP501 tra operon in the DNA secretion process is indicated. PG, peptidoglycan; CM, cytoplasmic membrane; CP, cytoplasm.
