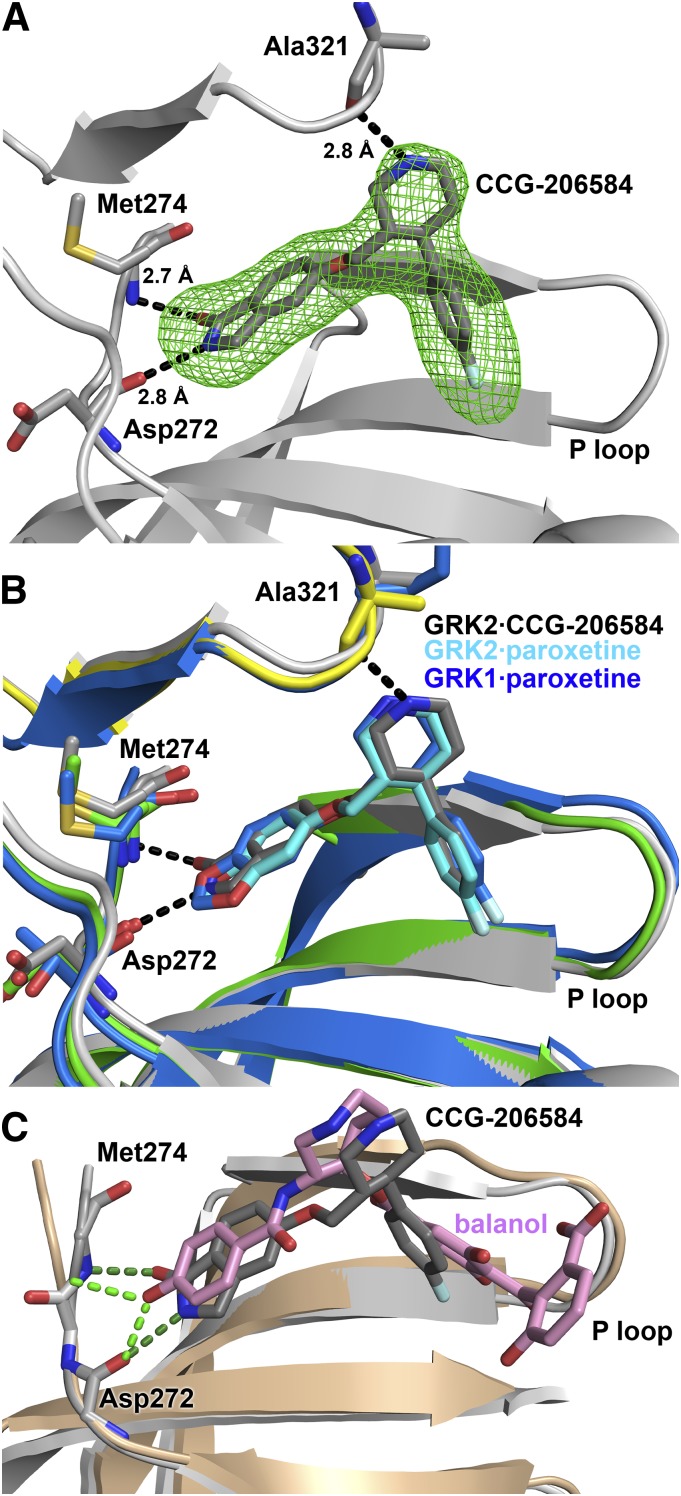
Fig. 7.
Crystallographic analysis of the GRK2·CCG-206584 complex. (A) Electron density Fo-Fc omit map contoured at 3σ (green mesh) of CCG-206584 bound to the active site of GRK2 (gray cartoon). The compound makes specific hydrogen bonds to backbone atoms of Asp272, Met274, and Ala321, shown as black dashed lines. (B) Superposition of the active sites of the GRK2·paroxetine–Gβγ, GRK2·CCG-206584–Gβγ, and GRK1·paroxetine complexes based on alignment of their small lobes. The small and large lobes of the kinase domain in the GRK2·paroxetine complex are colored green and yellow, respectively, and the carbons of paroxetine are colored cyan. The GRK1 complex is colored blue, with blue carbons in paroxetine. All molecules bind in a similar conformation in the active site. The rationally designed CCG-206584 undergoes a small rotation toward the hinge region of GRK2 that allows it to form 2.7 and 2.8 Å hydrogen bonds with Asp272 and Met274, respectively. (C) Superposition of the GRK2·balanol complex (PDB ID 3KRW) with that of GRK2·CCG-206584–Gβγ by pair fitting key Cα residues in the active site (GRK2 residues 201, 272, 273, 274, and 321). GRK2 in complex with balanol is drawn in the color beige. Despite distinct relative conformations adopted by the large and small lobes of each kinase, both CCG-206584 (dark gray carbons) and balanol (pink carbons) form analogous hydrogen bonds (balanol green and CCG-206584 dark green) to backbone atoms in the hinge.