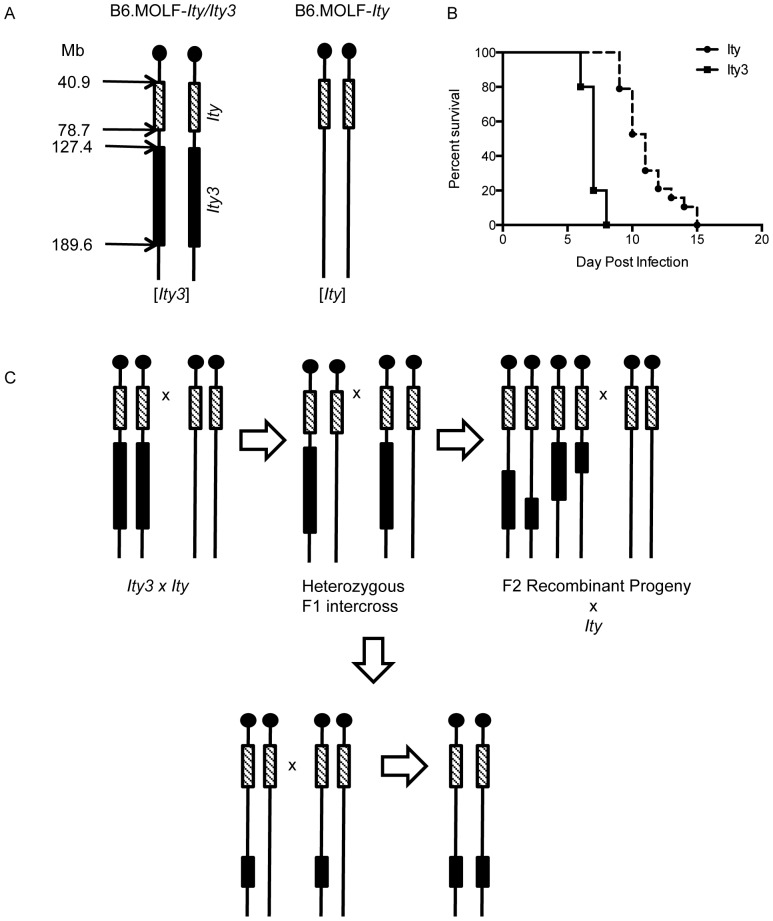
Figure 1. Breeding scheme used to generate the sub-congenic mice.
A) Schematic representation of mouse chromosome 1 illustrating the position of the congenic intervals Ity and Ity3 in B6.MOLF-Ity/Ity3 and B6.MOLF-Ity mice that were used to generate the subcongenic strains. The position of the congenic intervals is shown in megabases (Mb) according to Ensembl mouse genome Browser. (B) Survival curves of parental B6.MOLF-Ity/Ity3 (Ity3) and B6.MOLF-Ity (Ity) mice. Ity3 congenic mice are more susceptible to Salmonella infection, with a Log-Rank (Mantel-Cox) p-value <0.0001. Using the parental Ity and Ity3 congenic strains, subcongenic mice were generated using the breeding scheme shown in (C). The Ity and Ity3 mice were intercrossed to generate heterozygous mice, which were further backcrossed to the parental Ity mice. Using marker-assisted breeding, 12 subcongenic strains were generated. Boxes represent the MOLF/Ei allele, while the solid black lines represent the C57BL/6J allele. The shaded boxes represent the MOLF/Ei allele at the Ity locus and solid black boxes represent the MOLF/Ei allele at the Ity3 locus.