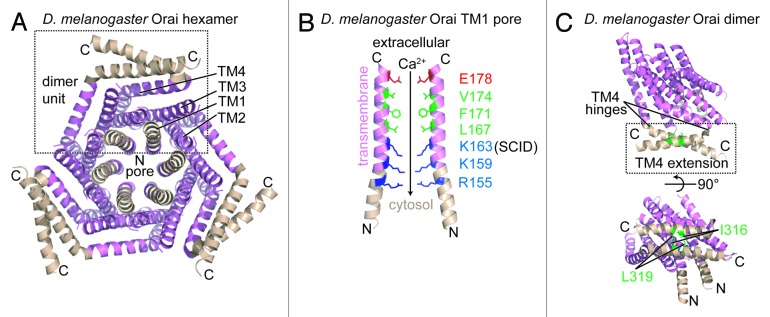
Figure 4.D. melanogaster Orai channel structure. (A) Cytosolic view of the Orai hexamer structure in the presumably closed state. An individual dimer unit building block is bounded by a broken black box. The TM1, TM2, TM3 and TM4 segments from a single monomer are labeled. (B) Residue composition of the TM1-consituted pore region. Only 2 TM1 segments exhibiting the greatest separation are shown for clarity. The acidic (red), hydrophobic (green), and basic (blue) pore-lining residues are shown relative to the extracellular space and the cytosol. The residue position mutated in a heritable form of severe combined immunodeficiency disease (i.e., R91W in human numbering) is labeled as ‘SCID’. The direction of the Ca2+ gradient (i.e., high to low concentration) is indicated with an arrow. (C) The TM4 C-terminal extension within the dimer unit. The antiparallel interaction between the C-terminal extensions is shown with hydrophobic residues involved in stabilizing this dimer interface depicted as sticks (green). The hinge regions responsible for creating the antiparallel orientation of the C-terminal extensions are indicated. In (A–C), color is consistent with Figure 2, and the amino and carboxy termini are labeled N and C, respectively. The D. melanogaster structure images were created with the 4HKR.pdb coordinate file.51
