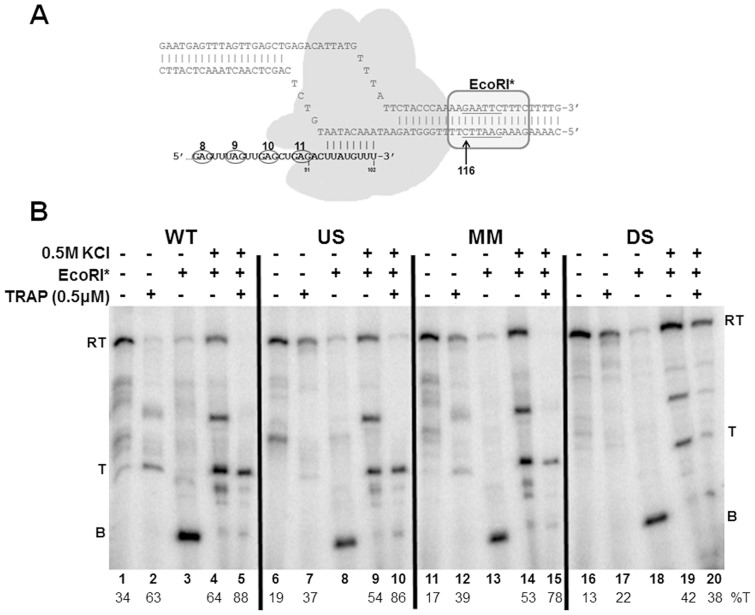
Figure 7. Blocking transcription elongation with EcoRI* to provide time for TRAP binding.
(A) Diagram of EcoRI* blocking the transcription elongation complex. Cleavage defective E111Q EcoRI (EcoRI*) binds to its recognition site on the DNA template and blocks elongation of RNAP approximately 12–13 bp upstream of the first G of the GAATTC recognition site. EcoRI* is shown as an oval shape bound to its recognition site starting at +116 in the trp leader region. RNAP is shown as a shaded grey shape. The nascent RNA is shown in bold with the 3′-most 8 residues paired with the template DNA. The last 4 (G/U)AG repeats of the TRAP binding site are circled. (B) Gel electrophoresis using a 6% polyacrylamide-8M urea gel analysis of block-and-release assay for the wild type (WT), U-stretch Disruption (US), C125G mismatch stem (MM), and disrupted stem (DS) attenuator templates. EcoRI* was allowed to bind to the DNA template prior to initiating transcription, TRAP was added, and transcription was then allowed to proceed until the TECs were blocked by EcoRI*. EcoRI* was then dissociated from the DNA by addition of 0.5 M KCl, allowing transcription to resume. The location of transcripts from blocked TECs (B), terminated at the attenuator (T), and read through (RT) transcripts are indicated on both sides of the gel. The percentage of transcription termination is displayed below the lane numbers (%T).