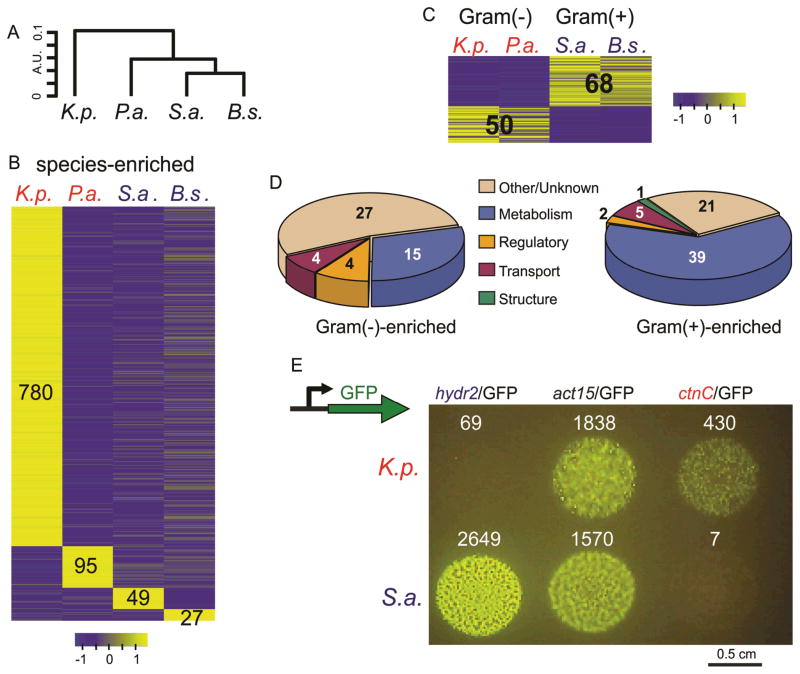
Figure 1. Changes in the physiological response in D. discoideum when feeding on different bacteria.
a, A dendrogram depicting the distances between the transcriptomes of D. discoideum growing on K. pneumoniae (K.p.), P. aeruginosa (P.a.), S. aureus (S.a.), and B. subtilis (B.s.). The dendrogram was constructed by hierarchical clustering (R function hclust) on the average normalized expression vectors of the two biological replicates from each growth condition consisting of all the genes from RNAseq experiments. We used Spearman’s correlation (SC) to calculate the distances (D = 1-SC) and complete linkage as the clustering criterion. Two objects (individual transcriptomes or joints) are joined by means of a horizontal line if these objects are more similar to one another than any other object in the data. The vertical distance between objects is inversely proportional to the similarity between them, but the horizontal distances are meaningless. b, c, The heat maps represent the patterns of change in standardized mRNA abundance for genes that were differentially expressed in D. discoideum when grown on different bacterial species. Each row represents a gene and each column, a bacterial growth condition. The colors represent relative mRNA abundances. To allow for comparisons between gene expression profiles with different abundances, we normalized the measurements on each gene to have a mean of 0 and a standard deviation of 1, and the scale indicates the number of standard deviations that a measurement is above or below the mean. b, Heat maps representing genes that are differentially expressed between the 4 bacterial species (species-enriched genes). c, genes differentially expressed between D. discoideum cells grown on Gram(+) and Gram(−) bacteria. d, pie charts showing the proportion of group-enriched D. discoideum genes categorized by Gene Ontology annotation (biological process). e, Wild-type amoebae transformed with a green fluorescent protein (GFP) coding sequence placed under the control of D. discoideum bacterial responsive gene promoters, mixed with different bacteria and spotted on buffered agar. A D. discoideum strain expressing GFP under the control of the hydr2 promoter fluoresces specifically when exposed to S.a., a Gram(+) bacterium, but not on K.p., a Gram(−) bacterium, and D. discoideum strain expressing GFP under the control of the ctnC promoter fluoresces when grown on K.p., but not on S.a.. A D. discoideum strain expressing GFP under the control of the actin15 promoter fluoresces under both growth conditions. Numbers indicate averaged normalized read counts from the RNA-seq data (supplemental Table S1).