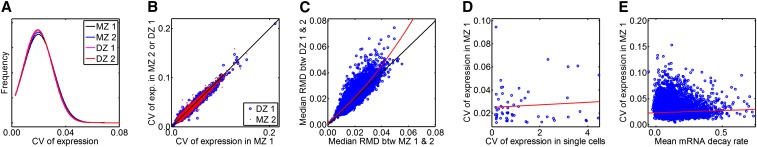
Figure 1.
Distributions of expression variability in LCLs. (A) Distribution of CVs of gene expression (probe n = 35,078) measured in MZ and DZ twins. MZ 1 is the set of first pairs of all MZ twins and MZ 2 is the set of second pairs of all MZ twins. Similarly, DZ 1 is the set of first pairs of all DZ twins and DZ 2 is the set of second pairs of all DZ twins. (B) Scatter plot of CVs of gene expression (probe n = 35,078) in MZ 1 against those in MZ 2 (blue) or DZ 1 (red) cohorts. (C) Scatter plot of median RMD between pairs of MZ twins against median RMD between pairs of DZ twins. Each blue dot indicates a single expression probe (or a gene) and the position of the blue dot indicates the median value of RMD of expression between all MZ pairs (MZ 1 − MZ 2) on the x-axis and that between all DZ pairs (DZ 1 − DZ 2) on the y-axis. The red line is based on quadratic regression to show a more pronounced difference between MZ and DZ with greater RMD. (D) Scatter plot of CVs of gene expression (n = 59) in single cells against CVs of gene expression in MZ 1. (E) Scatter plot of mean mRNA decay rate against CVs of gene expression in the MZ 1 cohort. The red line is based on the linear regression.