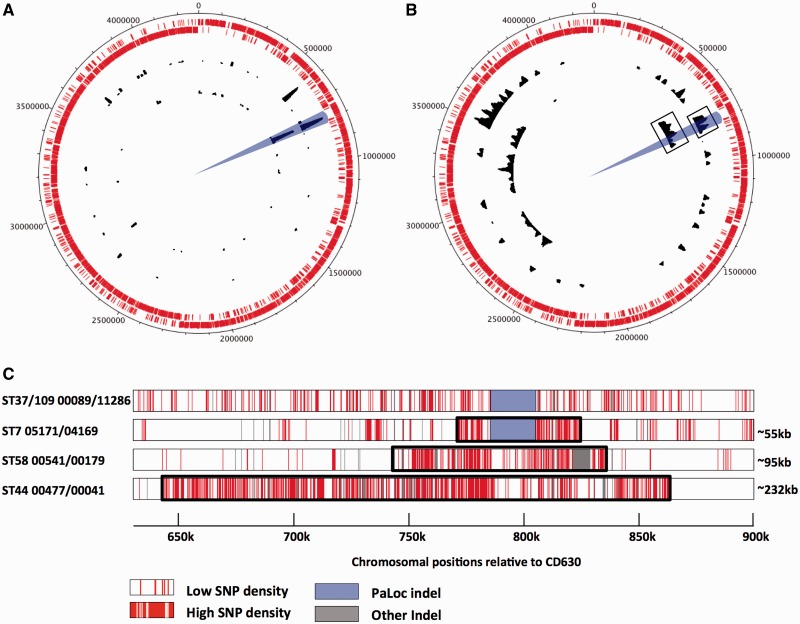
Fig. 4.—
PaLoc acquisition, loss, and exchange by homologous recombination involving long fragments of chromosomal DNA. (A) PaLoc acquisition and loss. Whole-genome distributions of indels between the two pairs of isolates marked by boxes in figure 3A (toxigenic ST37 and nontoxigenic ST109 outer black ring) and 3B (toxigenic and nontoxigenic ST7 inner black ring). The location of the PaLoc is indicated by blue shading. The two outer rings composed of small red lines indicate the open reading frames annotated on the forward and reverse strands of reference genome CD630 (Sebaihia et al. 2006). (B) PaLoc exchange within clade 1. Whole-genome distributions of polymorphism between the two pairs of isolates marked by boxes in figure 3C (toxigenic ST58, outer black ring) and 3D (toxigenic ST44, inner black ring). (C) Distribution of polymorphism between the four pairs of genomes shown in (A) and (B) within the region of the genome containing the PaLoc. Each row represents a pairwise comparison, and polymorphisms are shown in red. Enlarging the region of the genome flanking the PaLoc in this way allowed the distribution of polymorphisms to be used to estimate the size of the recombination events (black boxes) as ∼55 kb replaced by ∼36 kb during PaLoc loss by ST7, and ∼95 kb or ∼232 kb during PaLoc exchange within ST58 and ST44.