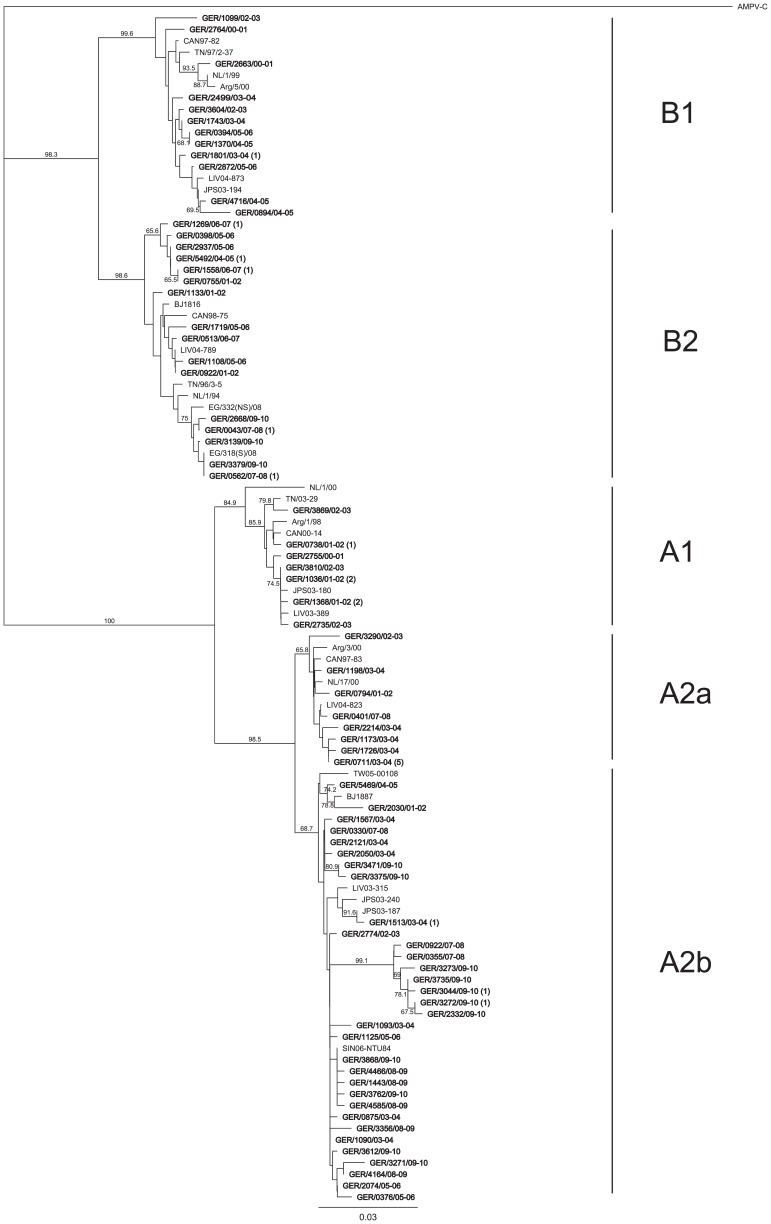
Figure 1. Phylogenetic tree of partial F gene fragments of German HMPV.
The tree was constructed using the maximum likelihood estimation with 1,000 replicates through Geneious 5.0.4. Avian metapneumovirus C (AMPV C) was included in the analysis and used as outgroup. Reference sequences (supporting information Table S1) representing the different HMPV genetic lineages were additionally included in the analysis. German HMPV sequences are shown in boldface. Nomenclature for German sequences uses a letter code representing the country of detection (e.g., “GER” represents Germany) followed by the patient number and season in which the virus was detected. In the case of identical sequences only one sequence is shown, whereas the number in brackets indicates the number of additional identical sequences. The lineages and sub-clusters are indicated to the right of the figure. Only bootstrap values greater than 65% are displayed at the branch nodes.