Figure 1. Multiple functions of BIR.
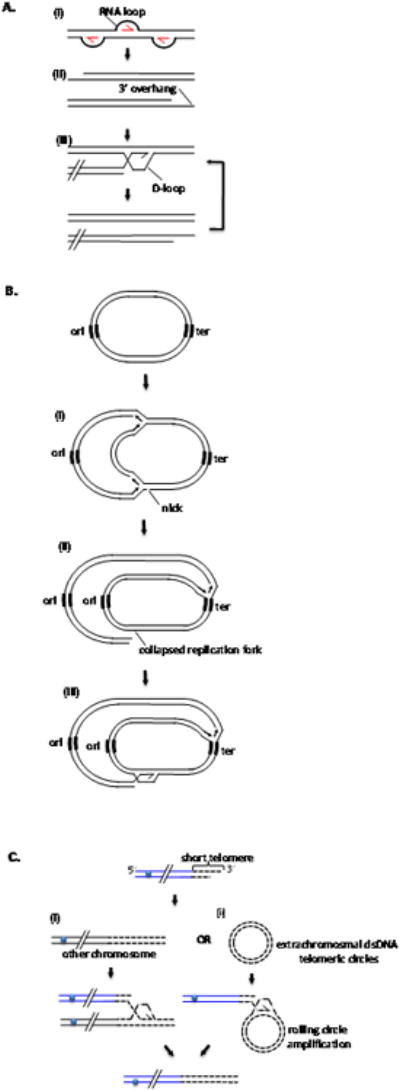
A. Replication of T4 bacteriophage. (i) Several R-loop structures mediate initial rounds of DNA replication. (ii) End-replication problem of lagging strand, collapsed replication forks or resection leave 3′ overhangs that initiate rounds of recombination mediated replication (RDR). (iii) Strand invasion occurring at homologous sequence present at the other end of the same molecule (this is possible because T4 chromosome is terminally redundant; not shown) or at internal part of co-infected molecule (shown) as T4 DNA is circularly permuted. B. Repair of broken replication forks in bacteria. (i) Replication fork runs into a nick and collapses (ii). (iii) Repair of collapsed fork by BIR. Since there is only one origin of replication (ori) extensive synthesis has to be primed from repaired replication fork to terminator (ter). C. Telomere lengthening by recombination. (i) BIR proceeds between telomeres of different chromosomes. (ii) BIR by rolling circle mechanism with extrachromosomal dsDNA telomeric circles.
