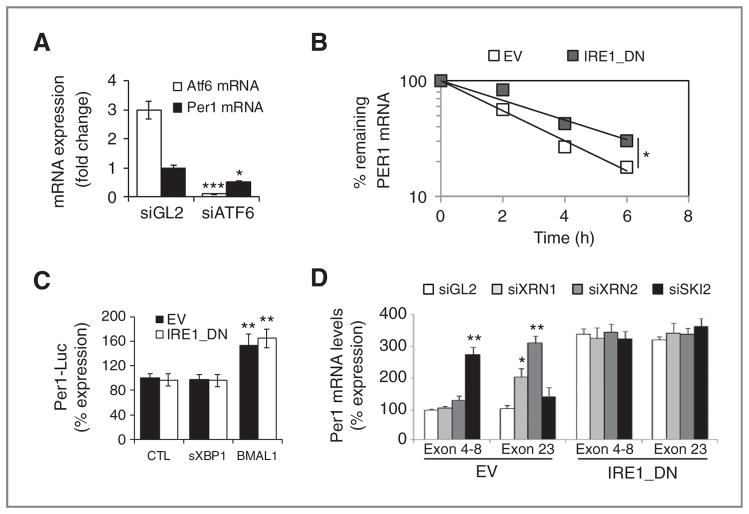
Figure 2.
IRE1α-mediated posttranscriptional control of PER1 mRNA in cultured cells. A, PER1 (closed) and ATF6 (open) mRNA expression as determined by quantitative RT-PCR in cells transfected with siRNA against luciferase (siGL2) and ATF6 (siATF6). Experiments were carried out in triplicate and the mean ± SD, statistical significance (Student t test) is indicated (*, P < 0.05; ***, P < 0.01). B, empty vector and IRE1_DN cells were cotransfected with control plasmid (pCMV-rL) or PER1 promoter-dependent luciferase reporter and either an empty pCDNA3 vector, a pCDNA3-sXBP1 vector, or a pCDNA3-BMAL1 vector. Cells were then lysed and lysates analyzed with the Dual-Luciferase Reporter Kit (Promega). Results were normalized against pCMV-Renilla luciferase (t test, **P < 0.05). C, actinomycin D pulse-chase was carried out as described in Materials and Methods. Total mRNA was extracted and quantitative RT-PCR experiments were conducted using PER1 mRNA-specific primer pairs. The experiment was repeated 3 times and data are presented as mean ± SD. Statistical significance was determined using Student t test, *, P < 0.03. D, empty vector and IRE1_DN cells were transfected with siRNA against XRN1/2 or SKI2. RNA was isolated after 48 hours and was used to amplify different regions of PER1 mRNA. Experiments were carried out in triplicate and the mean ± SD, statistical significance (Student t test) is indicated (*, P < 0.05; **, P < 0.01).