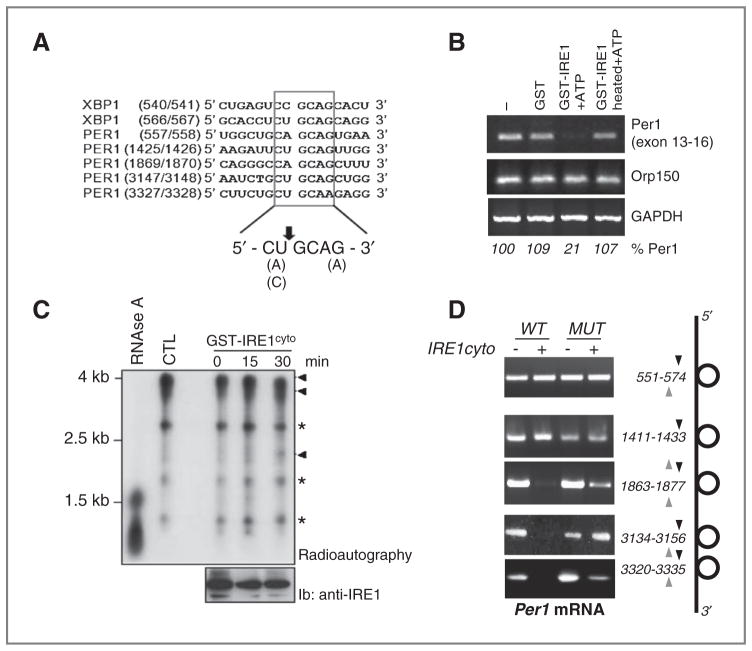
Figure 3.
IRE1α-mediated posttranscriptional control of PER1 mRNA in vitro. A, sequence alignment of XBP1 mRNA IRE1α-mediated cleavage sites with similar regions in PER1 mRNAs. B, in vitro RNA cleavage assay. Total RNA extracted from U87 cells was incubated with GST or GST-IRE1α-cyto in the presence of ATP for 2 hours at 37°C. RT-PCR was then conducted to determine PER1, ORP150, and GAPDH mRNA levels. C, PER1 cDNA sequence cloned into the pCDNA3 vector was used as template for in vitro transcription using the T7 Ribomax kit (Promega) in the presence of 32P-UTP. The resulting radiolabeled riboprobe was then incubated or not with dephosphorylated GST-hIRE1cyto for the indicated periods of time or with RNase A for 15 minutes at room temperature. The reaction products were resolved by PAGE and revealed by radioautography on X-ray films. The amount of recombinant GST-IRE1cyto added to the reaction is shown in the bottom blot using immunoblot with anti-IRE1 antibodies. *, nonspecific bands; Arrowheads, full and cleaved PER1 mRNA products. D, PER1 mRNA wild-type and mutated on each potential IRE1α cleavage sites were transcribed in vitro and subjected to in vitro cleavage with GST-hIRE1cyto as in F. Reaction products were then subjected to RT-PCR with specific primers flanking each cleavage site.