Table 1.
Model parameters and variable definitions. Variables can change during a simulation. Numerical values are parameters fixed for the duration of a simulation. Range values are deterministically calculated from other variables/parameters.
| symbol | description | value | unit |
|---|---|---|---|
| R | resource concentration | variable | μg ml−1 |
| Ni | density of host strain i | variable | cells ml−1 |
| Vj | density of phage strain j | variable | virions ml−1 |
| Ninit | initial host density | 4.6×104 | cells ml−1 |
| Vinit | initial phage density | 8.1×105 | virions ml−1 |
| ω | chemostat dilution rate | 0.0033 | min−1 |
| R0 | resource supply concentration | 2.2 | μg ml−1 |
| ɛ | resource conversion rate | 2.6×10−6 | μg cell−1 |
| γ | maximum resource uptake rate | 0.0123 | μg min−1 |
| K | half-saturation constant | 4 | μg ml−1 |
| δi | growth scaling for host hi | range [δmin,δmax] | scalar |
| δmin | minimum growth scaling factor | 0.8 | scalar |
| δmax | maximum growth scaling factor | 1.2 | scalar |
| ϕ | maximum adsorption rate | 0.104×10−8 | ml(min virion)−1 |
| θij | adsorption scaling for vj on hi | range [0,ϕ] | scalar |
| β | burst size | 71 | virions cell−1 |
| hi | genotype of bacteria i | range [0,1] | scalar |
| vj | genotype of phage j | range [0,1] | scalar |
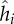 |
resistance phenotype of bacteria i | range [0,1] | scalar |
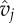 |
infection phenotype of phage j | range [0,1] | scalar |
| hinit | initial bacteria genotype | 0.2 | scalar |
| vinit | initial phage genotype | 0.2 | scalar |
| S | specificity of phage | 100 | scalar |
| μN | host mutation rate | 10−6 | cell−1 |
| μV | phage mutation rate | 10−5 | virion−1 |
| σN | s.d. of host mutation range | 0.01 | scalar |
| σV | s.d. of phage mutation range | 0.01 | scalar |
| MN | bacterial mutation size | random variable | scalar |
| MV | phage mutation size | random variable | scalar |
| Δt | integration time step | 10 | min |
| T | simulation duration | 5×107 | min |
| ρ | resolution of genotype diversity | 0.001 | scalar |
| L | chemostat volume | 1 | ml |
