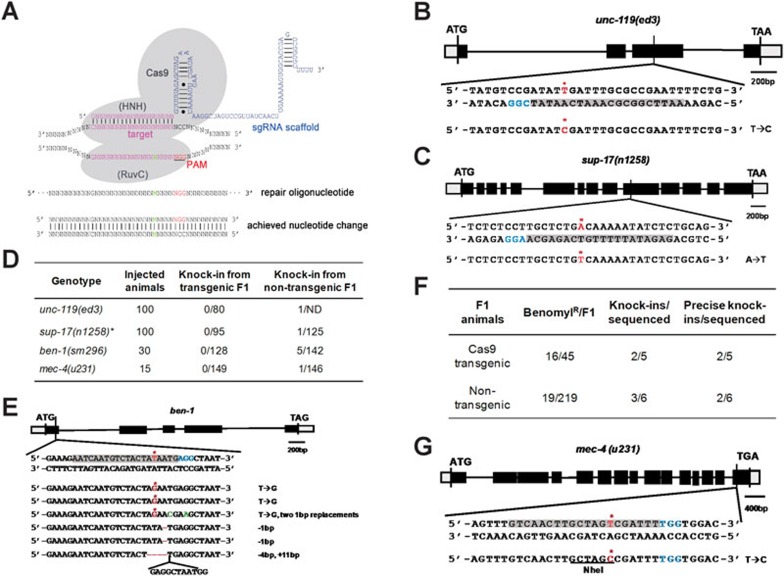
Figure 1.
Oligonucleotide-based gene editing in C. elegans. (A) A schematic diagram of the CRISPR/Cas9 system using an oligonucleotide as the repair template. sgRNA directs Cas9 to the target site where Cas9 cleaves the DNA sequence preceding the PAM sequence. The oligonucleotide, containing a single nucleotide change (indicated as M), serves as a template to repair the DSDB via HR. (B, C, G) Gene editing at the unc-119(ed3), sup-17(n1258) and mec-4(u231) loci. The sense or anti-sense DNA sequences targeted by sgRNAs are shaded in gray, and the PAM sequences in blue. The nucleotide targeted for substitution is in red and also marked with a red asterisk. The NheI site generated in G is underlined. (D) Summary of gene-editing experiments at 4 different genetic loci. *: a phosphorothioate-modified oligo was used as a repair template. ND: not determined. (E) Gene editing at the ben-1 locus. The sense-strand sequence targeted by sgRNA is shaded in gray. The nucleotide targeted for substitution is labeled in red and marked with a red asterisk. DNA sequences in the target region from homozygous benomyl-resistant progeny derived from non-transgenic F1 animals are aligned with the wild-type DNA sequence, with unanticipated nucleotide substitutions (green), deletions, and insertion indicated. (F) Summary of ben-1 gene editing. The numbers of benomyl-resistant (benomylR) F1 animals versus the number of Cas9 transgenic or non-transgenic F1 animals are shown. The number of F1 animals with the desired Amber mutation (knock-ins) and the number of F1 animals with the correct Amber mutation and no other mutations in the ben-1 gene (precise knock-ins) versus the number of benomylR F1 animals sequenced are also shown.