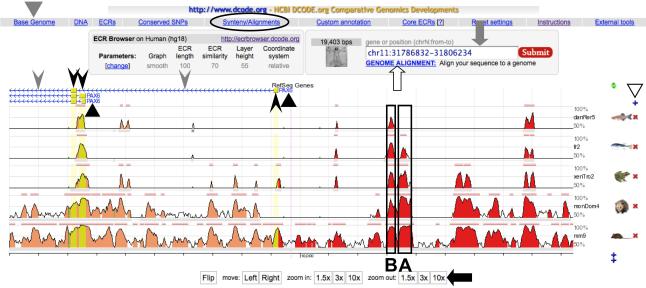
Fig. 1.
A screen shot of the ECR Browser window. Above the schematic representation of the alignment, the gene names (PAX6, indicated by black triangles), exons (boxes indicated by black arrowheads) and introns (horizontal lines indicated by gray arrowheads) in the base genome are shown. Arrows associated with the horizontal line representing introns indicate transcriptional orientation of the gene. The conserved regions shown as peaks include not only exons but also CNEs located in introns and intergenic regions. Among these CNEs, those indicated as A and B (boxed) were shown to have enhancer activity in the pancreas and lens/cornea in transgenic mouse analyses, respectively (32). One can change the base genome species by clicking the “Base Genome” button (indicated by a gray triangle) at the upper left, but it is not necessary to choose Xenopus at this step, even if it will be used as the base sequence in a subsequent PipMaker analysis. On the right can be seen a + or an × next to the genomes shown (white triangle), permitting addition or removal of genomes from the alignment. The length of the alignment is adjusted using buttons such as “10 × ” at the bottom of the window (black arrow), or by directly inputting a desired genomic position of the base genome into the rectangular box at the upper right (gray arrow). One may also input just a gene name into the rectangular box to move to another gene locus. If one needs to align user-identified sequence to the genomes in this browser, click the “GENOME ALIGNMENT” button below the rectangular box (white arrow), and input the sequence as a FASTA format. For details, go to the instruction page of ECR Browser (http://ecrbrowser.dcode.org/ecrInstructions/ecrInstructions.html).