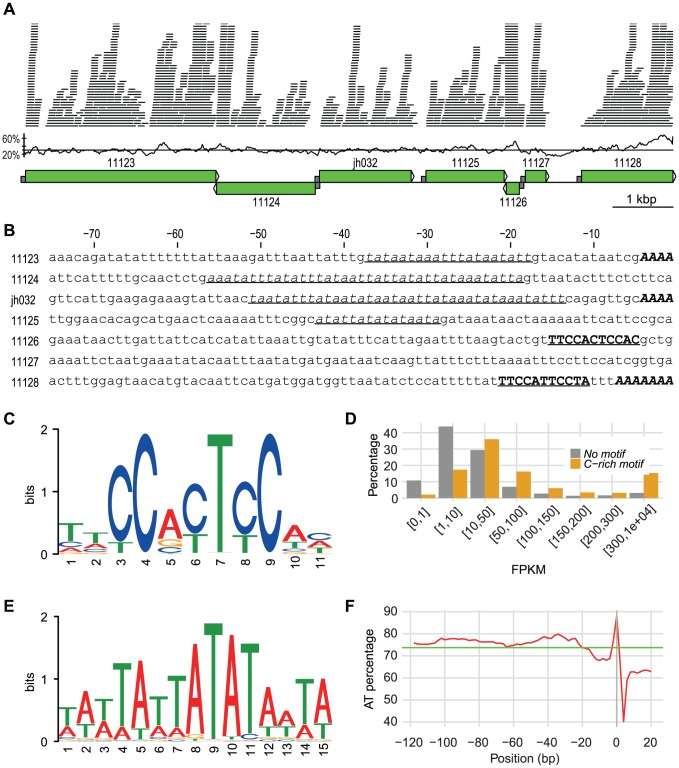
Figure 1. A 10 kbp genomic region with promoter info.
A 10 kbp long genomic region located on scaffold scf7180000020498. The first part shows the Illumina RNA-Seq reads mapped onto the region. A coverage cutoff 40 was used for a better display. The GC content in 100 bp windows with a step size of 20 bp is shown. The average GC content in the region (30.5%) is indicated by a line. Green boxes with arrows indicate position and direction of annotated genes. Half sized grey boxes indicate 75 bp promoter regions with sequences shown in B. Numbers refer to protein IDs: 11123, Phospholipid-transporting ATPase; 11124, Hypothetical protein; jh032, Hypothetical protein; 11125, Long-flagella protein, kinase, CMGC RCK; 11126, Ribosomal protein S30; 11127, Prefoldin subunit 6; 11128, ATP-dependent RNA helicase. B. 75 bp promoter sequences of the genes shown in A. TATA-box motifs are in underlined italic font, C-rich motifs are in bold underlined upper cases, and the multiple As before start codon are in bold italic upper cases. C. Sequence logo of C-rich motif. D.Comparison of expression between genes with and without C-rich motif. Genes are divided into eight categories based on their FPKM-values. The y-axis represents the percentage of genes within each category. E. Sequence logo of TATA-box motif. F. AT contents in percentage of the 20 bp C-terminus of the genes with their 120 bp promoter regions drawn with window size of 3 and step size of 2. X-axis shows the positions, while y-axis shows the AT percentage. Green line indicates the average AT percentage of the regions. Plot was drawn in R.