Abstract
Background
MicroRNAs (miRNAs) represent a class of small ncRNAs that repress gene expression on the post-transcriptional level by the degradation or translation inhibition of target mRNA.
Methodology
Three small RNA libraries from oyster haemocytes were sequenced on the Illumina platform to investigate the latent immunomodulation of miRNAs after bacteria challenge and heat stress. Totally, 10,498,663, 8,588,606 and 9,679,663 high-quality reads were obtained in the control, bacteria and bacteria+heat library, respectively, from which 199 oyster miRNAs including 71 known and 128 novel ones were identified. Among these miRNAs, 6 known and 23 novel ones were predicted to possess more than one precursor-coding region, and cgi-miR-10a, cgi-miR-184b, cgi-miR-100, cgi-miR-1984 and cgi-miR-67a were observed to be the most abundant miRNAs in the control library. The expression levels of 22 miRNAs in the bacteria library were significantly higher than those in the control library, while there were another 33 miRNAs whose expression levels were significantly lower than that in the control library. Meanwhile, the expression levels of 65 miRNAs in the bacteria+heat library changed significantly compared to those in the bacteria library. The target genes of these differentially expressed miRNAs were annotated, and they fell in immune and stress-related GO terms including antioxidant, cell killing, death, immune system process, and response to stimulus. Furthermore, there were 42 differentially expressed miRNAs detected in both control/bacteria and bacteria/bacteria+heat comparisons, among which 9 miRNAs displayed the identical pattern in the two comparisons, and the expression alterations of 8 miRNAs were confirmed using quantitative RT-PCR.
Conclusions
These results indicated collectively that immune challenge could induce the expression of immune-related miRNAs, which might modulate the immune response such as redox reaction, phagocytosis and apoptosis, and the expression of some immune-related miRNAs could be also regulated by heat stress to improve the environmental adaption of oyster.
Introduction
MicroRNAs (miRNAs) are endogenously encoded single-stranded non-coding RNAs of about 22 nt in length [1]. They are initially transcribed by RNA polymerase II in the nucleus as primary miRNAs, which are cleaved by the nuclear RNase III type enzyme Drosha to produce a short hairpin precursor miRNA. After transferring into the cytoplasm, the precursor miRNA is further cleaved by Dicer into the functional double-stranded RNA, which is incorporated into the RNA-induced silencing complex (RISC) and forms the mature miRNA [2], [3]. So far, a large number of miRNAs have been identified in various metazoans, many of which are evolutionarily conserved, and have evolved to be potent regulators of gene expression on the post-transcriptional level [4].
Mature miRNAs have the ability to regulate the stability and/or translational efficiency of their mRNA targets in metazoa through the imperfect base-pairing between target transcript and the 5′ seed region of the miRNA [5]. It has been reported that more than 60% of mammalian protein-coding genes are computationally predicted as targets of miRNA [6]. In addition, it has been considered that one gene can contain multiple miRNA binding sites, and one miRNA can regulate hundreds of target mRNAs, resulting in a complex gene-regulatory network to implement the spatio-temporal coordination of gene expression under specific development stage or physiological status [1], [5], [7]. The miRNA-coordinated gene expression contributes to the maintenance of homeostasis and the improvement of host adaption [8].
As a regulator of gene expression on the post-transcriptional level, miRNAs play an important role in the modulation of many biological processes to confer robustness on these biological processes, and further maintain the tissue identity in a variety of metazoans [9]. It has been evidenced that miRNAs are able to modulate host immune and stress responses [8], [10]–[13]. The expression of immune-related miRNAs in immunocytes can be regulated by the immune response against the invasive pathogens [14], and then these miRNAs can modulate properly the expression of pattern recognition receptors, signal pathway molecules or immune transcription factor to regulate the host-pathogen interaction and the elimination of invasive pathogens [15]–[18]. For example, mammalian NF-κB signal pathway is modulated dynamically by a set of miRNAs (including let-7, miR-9, miR-21, miR-218, and so on) during the whole process of immune response [16], [19]. In addition, stress response can also alter the biogenesis of miRNAs [20], and then the miRNAs function as the buffer to attenuate the harmful effect of stresses on some physiological activities [11], [21]. Mollusca are a large and diverse phylum in invertebrates, and there are several reports about the identification of miRNAs in some species. To our knowledge, there are 5 miRNAs identified from Haliotis rufescens and 60 miRNAs from Lottia gigantean [22]. However, there is still no any report about the physiological regulation function of mollusc miRNAs, specially their immunomodulation.
Because of the economic and ecological importance of Pacific oyster Crassostrea gigas, it has become a suitable model organism for studying immune and stress response in marine bivalves [23], [24]. Furthermore, the recent released genome sequence provides the relatively complete genetic information for the exploration of physiological function of oyster miRNAs [25]. Investigations of miRNAs in oyster C. gigas will pave a new way to further understand the modulation mechanism of immune-related genes in molluscs during the immune and stress response. The purposes of this study were to (1) identify the known and novel miRNAs from oyster C. gigas, (2) survey the expression alteration of all identified miRNAs in haemocytes of oyster after bacteria challenge and heat stress, (3) predict the target genes of miRNAs, and analyze GO information of the target genes of differentially expressed miRNAs to understand the potential immunomodulation of miRNAs in oyster.
Result
Overview of Small RNA Library Sequencing
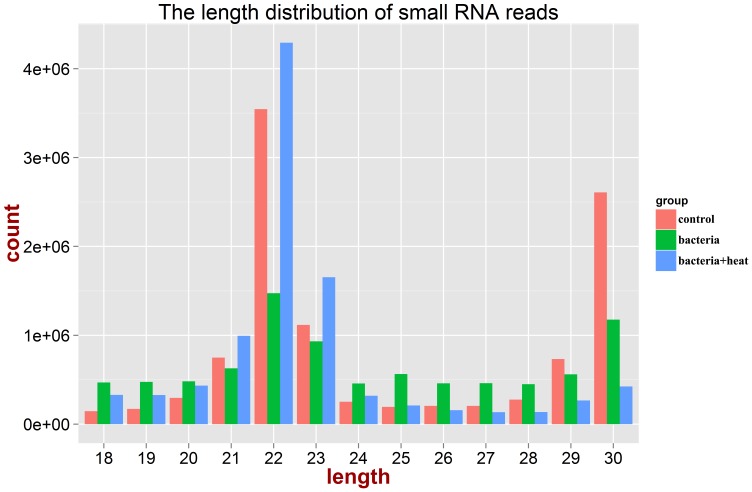
The small RNA libraries from the haemocyte samples in the control, bacteria and bacteria+heat groups were sequenced by Illumina deep sequencing technology to survey the miRNA transcriptome in oyster. Totally 11,760,772 raw reads were obtained from the control library, 11,361,851 from the bacteria library, and 12,165,062 from the bacteria+heat library. After the discarding of low-quality sequence, adaptor sequence and sequences shorter than 18 nt and longer than 30 nt, 10,498,663, 8,588,606 and 9,679,663 high-quality reads were remained for the statistics analysis of sequence length (Table 1), and 51.6%, 35.3% and 71.7% reads were of 21∼23 nt in the control, bacteria and bacteria+heat library, respectively (Fig. 1).
Table 1. Statistics for the distribution of reads filtered in order.
| Reads in the control group | Reads in the bacteira group | Reads in the bacteria+heat group | |
| Total | 11760772 | 11361851 | 12165062 |
| Low quality filter | 317263 (2.7%) | 427064 (3.8%) | 339547 (2.8%) |
| Adaptors filter | 68014 (0.6%) | 30211 (0.3%) | 51019 (0.5%) |
| Length filter | 876832 (7.5%) | 2315970 (20.4%) | 2094833 (17.2%) |
| Remaining reads | 10498663 | 8588606 | 9679663 |
Figure 1. Length distribution of high-quality reads in the three libraries.
Red: control library; Green: bacteria library; Blue: bacteria+heat library.
After the merging of reads from three libraries and the removal of redundancy, 229,302 clean sequences with more than 3 reads were retained for alignment analysis. After comparing the clean RNA sequences with the Rfam databases, Repbase databases and oyster mRNAs, a total of 80,005 sequences derived from other non-coding RNA, repeat sequence or mRNA degradation product were removed (Table 2). The remaining 149,297 sequences were retained for further miRNA identification analysis.
Table 2. Statistics for the filtered clean reads.
| Remaining Reads | ||
| Total | 28766932 | |
| Remove redundancy | 2345828 | |
| Less than 3 reads | 229302 | |
| Hit Rfam and Repbase | 194881 | |
| Hit mRNA | 149297 | |
| Mapped miRNA | 2292 | |
The Discovery Of known and Novel miRNAs in Oyster
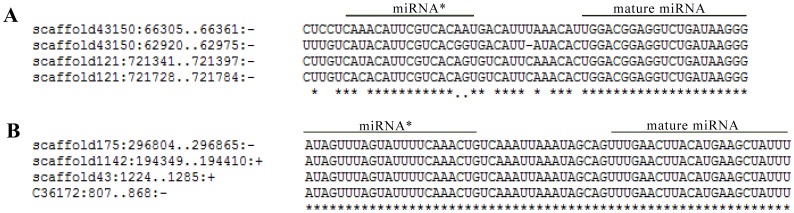
To identify known miRNAs in oyster, 149,297 filtered unique sequences were aligned against mature miRNAs in miRBase (version 19), and 2292 sequences homologous to registered mature miRNAs (with not more than one mismatch between sequences) were obtained. These homologous sequences were mapped to oyster genome sequence and further parsed through the miRDeep2 software for the prediction of precursor sequence and secondary structure. A total of 71 known miRNAs were identified in oyster (Table S1) with copy numbers ranging from 0 (cgi-miR-242) to 1,126,609 (cgi-miR-10a) in the control library (Table S3). The sequences that did not match registered mature miRNAs were also aligned with the oyster genome sequence to discover potential novel miRNAs, and total of 128 novel miRNAs were identified by miRDeep2 software (Table S2) with copy numbers ranging from 0 (scaffold888_4140, scaffold347_3876, scaffold1827_3866, scaffold43878_3031, scaffold35684_4427, scaffold531_824, scaffold42948_2113, scaffold535_3052, scaffold1174_3059, scaffold1769_5047) to 38,193 (scaffold42648_5080) in the control library (Table S3). In total, 199 oyster miRNAs were identified including 71 known and 128 novel ones, and the nucleotide sequences and genome coding regions of their precursors were shown in Table S1 and S2. Among these 199 oyster miRNAs, 6 known and 23 novel ones were observed to have more than one precursor-coding region. Furthermore, there were four potential precursors in the oyster genome for cgi-miR-184a, cgi-miR-184d and scaffold175_3234 (Fig. 2).
Figure 2. Precursor sequence alignment of miRNAs having four potential coding regions.
(A) miR-184a; (B) scaffold175_3234.
Differentially Expressed miRNAs during Bacteria Challenge and Heat Stress
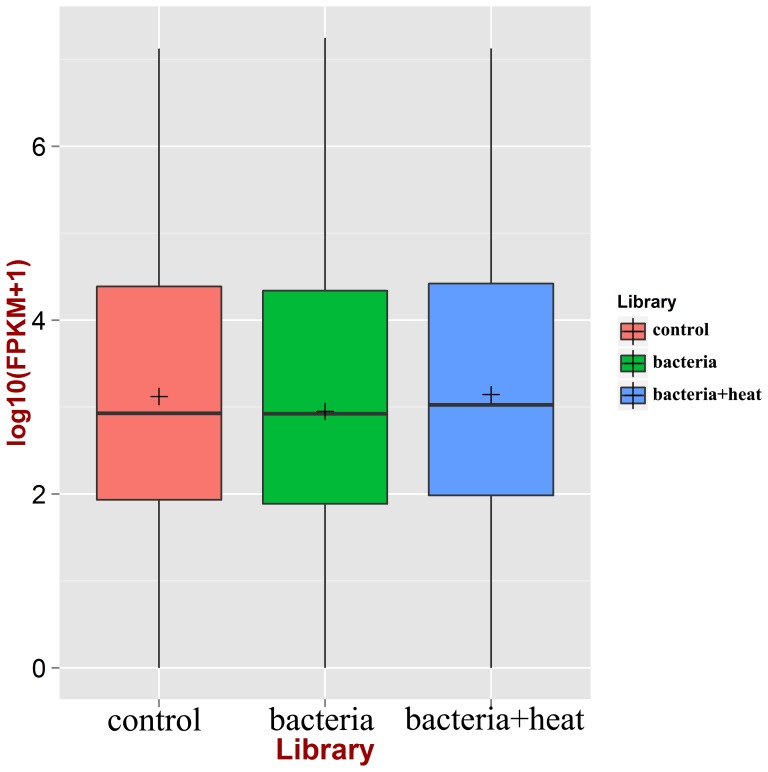
In the control library, 5 miRNAs including cgi-miR-10a, cgi-miR-184b, cgi-miR-100, cgi-miR-1984, cgi-miR-67a were identified with higher copy numbers, and their copy numbers were 1,126,609, 957,131, 308,405, 234,455 and 179,096, respectively. The copy numbers of oyster miRNAs were also counted in the control, bacteria and bacteria+heat library, respectively, and further converted to their expression levels in the form of FPKM (Table S3). The expression level of all oyster miRNAs in the three libraries was analyzed and shown in Fig. 3. The overall expression level of oyster miRNAs in the bacteria library was lower than that in the control group, while the level in the bacteria+heat library was higher than that in the control and bacteria libraries.
Figure 3. The overall expression level of all identified miRNAs in three libraries.
Red: control library; Green: bacteria library; Blue: bacteria+heat library. The overall expression level of oyster miRNAs in the bacteria+heat and bacteria library was highest and lowest, respectively.
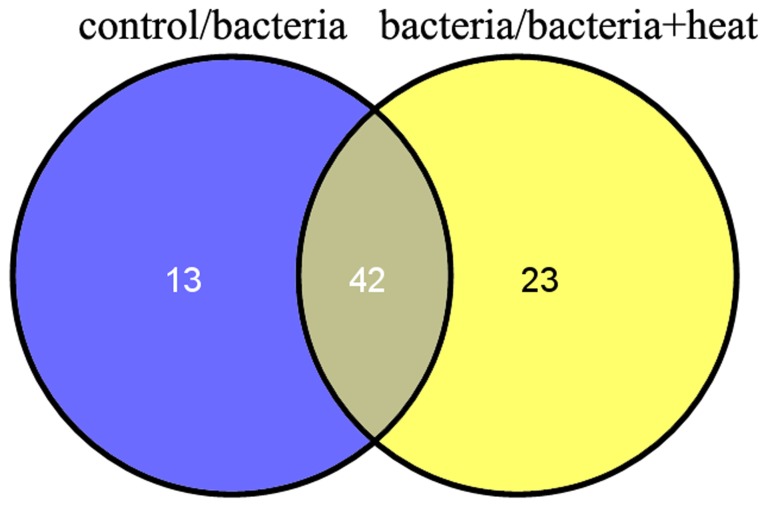
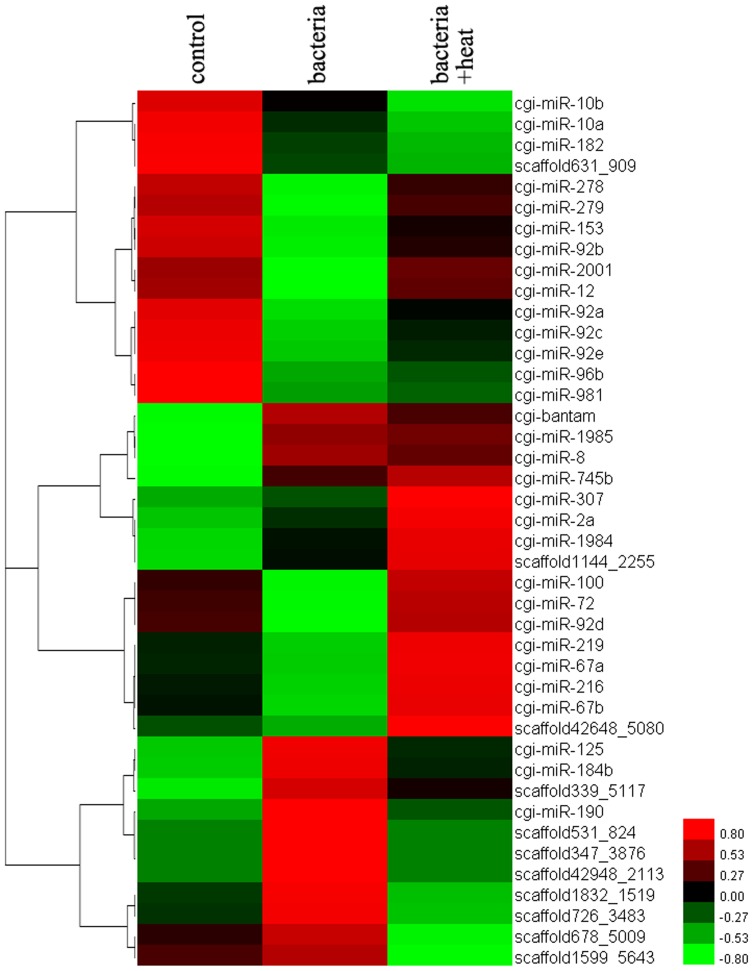
Based on the statistical analysis of copy numbers, the expression of 55 miRNAs in the bacteria library changed significantly compared to that in the control group, among which 22 and 33 ones increased and decreased significantly, respectively. Meanwhile, the expression levels of 65 miRNAs in the bacteria+heat library also changed significantly (37 miRNAs increased and 28 miRNAs decreased) compared to that in the bacteria library (Table S4). Furthermore, the expression of 42 miRNAs altered significantly in both control/bacteria and bacteria/bacteria+heat comparisons (Fig. 4). Among the 42 miRNAs, the expression level of 5 miRNAs (cgi-miR-2a, cgi-miR-307, cgi-miR-745b, cgi-miR-1984 and scaffold1144_225) increased significantly in both two comparisons, while the expression level of 4 miRNAs (cgi-miR-10a, cgi-miR-10b, cgi-miR-182 and scaffold631_909) decreased significantly (Fig. 5).
Figure 4. Grouping of differentially expressed miRNAs between two groups among the control, bacteria and bacteria+heat library.
Figure 5. Summary cluster map of miRNAs expression.
The miRNAs all changed significantly between two groups among the control, bacteria and bacteria+heat library. There was the identical alteration trend in the expression level of 9 miRNAs (cgi-miR-2a, cgi-miR-307, cgi-miR-745b, cgi-miR-1984, scaffold1144_2255, cgi-miR-10a, cgi-miR-10b, cgi-miR-182 and scaffold631_909) between the control/bacteria and bacteria/bacteria+heat comparisons. In addition, the alteration trend of other 33 miRNAs expression was contrary between the two comparisons.
The Confirmation of Differentially Expressed miRNAs
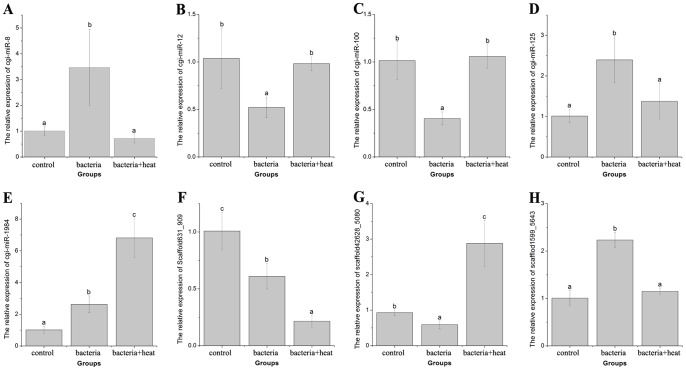
The expression of eight differentially expressed miRNAs in both control/bacteria and bacteria/bacteria+heat comparisons, including cgi-miR-8, cgi-miR-12, cgi-miR-100, cgi-miR-125, cgi-miR-1984, scaffold631_909, scaffold42648_5080 and scaffold1599_5643 (Fig. 6 A–H), were confirmed by quantitative real-time PCR. The expression alterations of the eight miRNAs were consistent between the small RNA sequencing and quantitative real-time PCR. The expression level of cgi-miR-8, cgi-miR-125, cgi-miR-1984 and scaffold1599_5643 increased significantly (P<0.05), whereas the expression level of other four miRNAs decreased significantly (P<0.05) in oyster haemocytes of the bacteria group, relative to that in the control group. Furthermore, cgi-miR-1984 expression level also increased significantly (P<0.05) in oyster haemocytes of the bacteria+heat group compared to that in the bacteria group (Fig. 6E), and scaffold631_909 expression level also decreased significantly (P<0.05) (Fig. 6F). However, the expression level of other six miRNAs was reverted in oyster haemocytes of the bacteria+heat group in comparison to that in the bacteria group.
Figure 6. The expression of eight miRNAs detected by SYBR Green real-time PCR in oyster haemocytes after bacteria challenge and heat stress, including cgi-miR-8 (A), cgi-miR-12 (B), cgi-miR-100 (C), cgi-miR-125 (D), cgi-miR-1984 (E), scaffold631_909 (F), scaffold42648_5080 (G) and scaffold1599_5643 (H).
5S gene was used as an internal control to calibrate the cDNA template for all the samples. Each values were shown as mean ± SD (N = 5), and bars with different letters were significantly different (P<0.05).
GO Analysis of Target Genes of Differently Expressed miRNAs
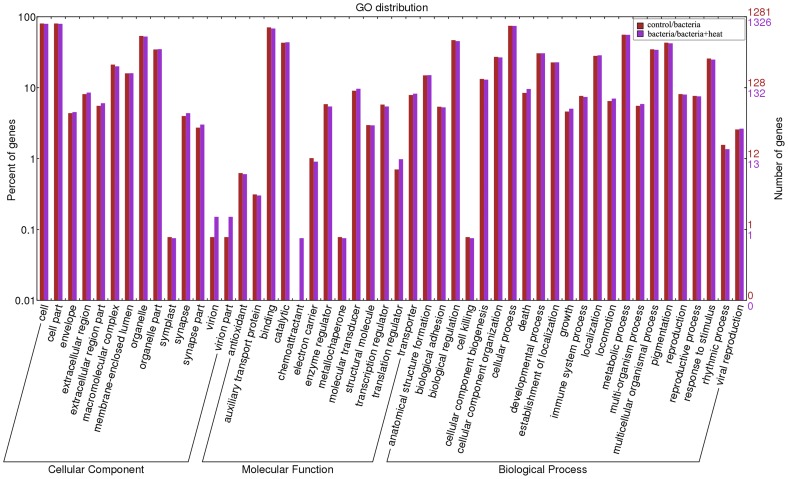
The target genes of all identified miRNAs were predicted using the miRanda algorithm, and there were total of 7696 pairing between protein-coding genes and miRNAs, which were equivalent to 5178 targets genes for 199 oyster miRNAs (Table S5). The target genes of 55 and 56 miRNAs were retrieved expressed differently in the control/bacteria and bacteria/bacteria+heat comparisons, respectively, and then assigned GO terms by Blast2GO software. The GO annotation of target genes were parsed and displayed through WEGO (Level-2) (Fig. 7). These GO annotation included 14 terms in Cellular Component ontology, 13 terms in Molecular Function ontology, and 23 terms in Biological Process ontology. Some terms (including antioxidant, cell killing, death, immune system process and response to stimulus) were correlated obviously with the physiological status of oyster exposure to bacteria challenge or heat stress. Furthermore, the term chemoattractant was observed in the comparison of the bacteria and bacteria+heat library, while not in the comparison of the control and bacteria library.
Figure 7. GO distribution of target genes of differentially expressed miRNAs.
Red: the target genes of differentially expressed miRNAs between the control and bacteria group; Purple: the target genes of differentially expressed miRNAs between the bacteria and bacteria+heat groups.
Discussion
MiRNAs represent a class of small non-coding RNAs that repress gene expression on a post-transcriptional level by the degradation or translation inhibition of target gene mRNA. It is well-known that miRNAs can modulate several physiologic and pathologic processes, including host immune response and stress response [26], [27]. In the present study, three small RNA libraries were prepared from haemocytes of oysters after bacteria challenge and heat stress, and they were sequenced on the Illumina Hiseq2000 platform to identify the miRNAs in oyster. After the filtering of low-quality sequence, adaptor sequence and sequences shorter than 18 nt and longer than 30 nt, a total of 28,766,932 high-quality reads were obtained and the most abundant reads were 22 nt ones in all three libraries. The length distribution of reads in the present study was consistent with the reports from other metazoans [28]–[30], demonstrating that three small RNA libraries from oyster were technically reliable and suitable for the subsequent analysis. After the removal of other non-coding RNA, repeat sequence and mRNA degradation product, totally 199 miRNAs were identified in oyster haemocytes, among which 71 ones were known and 128 ones were novel. The number of oyster miRNAs identified in the present study was more than that of other molluscs including H. rufescens and L. gigantean [22], and the obvious difference might result from the distinct sequencing depth of small RNA library. The 128 novel miRNAs might be of mollusc-specific, because they were not mappable to any reported mature miRNAs in miRBase databases. Among all the miRNAs, cgi-miR-10a, cgi-miR-184b, cgi-miR-100, cgi-miR-1984 and cgi-miR-67a were top 5 most abundant ones in the control library. The expression of miR-184 and miR-10 was also observed to be higher in Apostichopus japonicas and Plutella xylostella [28], [31], and their higher expression in oyster haemocytes suggested that they might play a crucial role in the maintenance of haemocyte physiological function [13]. Both miR-100 and miR-67 were considered to be ancient and conserved ones because they had been observed in almost all of metazoans [13], [32], but miR-1984 might be mollusc-specific because they were just identified in the other two molluscs. Interestingly, 29 miRNAs from oyster were observed to have more than one coding region of the precursors, implying that they could be transcribed from multiply sites and have more promoters, which might endow them more activation approaches at the transcription level under the multiple conditions and more chances to be involved in the modulation of physiological function [33]. The discovery of oyster miRNAs demonstrated that there were conserved and mollusc-specific miRNAs in oyster, and those 199 miRNAs expressed in oyster haemocytes might be involved in the modulation of host immune response and stress response.
MiRNAs modulation has been considered as a critical regulatory principle in the vertebrate immunity and adaptation [8], [34]. In order to understand the potential role of miRNAs in oyster immune response, the expression profile of 199 miRNAs in haemocytes was analyzed after bacteria challenge. The overall expression level of oyster miRNAs in the bacteria library decreased in comparison with that in the control group. Among all the identified miRNAs, the expression level of 22 miRNAs increased significantly after bacteria challenge, while the expression of 33 miRNAs decreased significantly. The results indicated that the 55 miRNAs in oyster haemocytes were of immune-responsive, and they might function as essential regulators of immune response in oyster [35], [36]. The significantly different expression of these miRNAs could result from the activation of miRNA biogenesis or degradation pathway triggered by the immune response against bacteria challenge in oyster haemocytes [3], [37]. The target genes of the 55 immune-responsive miRNAs were annotated by immune-related GO terms including antioxidant, cell killing, death and immune system process. These immune-responsive miRNAs could be implicated in the immunomodulation, and they might regulate the expression of target genes at the post-transcriptional level to modulate redox reaction, phagocytosis and apoptosis of oyster. The immune regulation of these miRNAs had also been reported in other animals [38], [39]. For example, miR-184 was observed to implicate in the modulation of apoptosis in some metazoans [40]. The present results suggested collectively that immune challenge could induce the expression of immune-related miRNAs in haemocytes, and these induced miRNAs might modulate the immune response via the regulation of redox reaction, phagocytosis and apoptosis in oyster.
To further explore the immunomodulation mechanism of oyster miRNAs under stress exposure, the expression level of miRNAs in haemocytes was evaluated after the simultaneous treatment of bacteria challenge and heat stress. The overall expression abundance of oyster miRNAs in the bacteria+heat library was highest among the three libraries. Meanwhile, the expression of 65 miRNAs in the bacteria+heat library changed significantly (37 miRNAs increased and 28 miRNAs decreased) in comparison with that in the bacteria library, similar to the 8 heat-responsive miRNAs from Brassica rapa [41]. Among the 65 miRNAs, 42 miRNAs also changed significantly in the expression levels in the control/bacteria comparison, indicating that heat stress response might modulate the immune response of oyster through the expression regulation of immune-related miRNAs. Furthermore, other 23 miRNAs which did not change significantly in the expression level in the control/bacteria comparison might represent heat-specific ones in oyster haemocytes. The 65 differentially expressed miRNAs in the bacteria/bacteria+heat comparison could be annotated the chemoattractant term in the Molecular Function ontology, which was not observed in the control/bacteria comparison. These results suggested that the expression level of immune-related miRNAs in haemocytes could be modulated by heat stress response, and heat-related miRNAs might modulate the expression of chemoattractant-related genes under heat stress to improve the heat adaption.
There were 42 miRNAs whose expression level changed significantly in both control/bacteria and bacteria/bacteria+heat comparisons. The expression levels of these miRNAs were clustered to understand their potent regulatory function during bacteria challenge and heat stress. Among the 42 miRNAs, the expression level of 5 miRNAs (cgi-miR-2a, cgi-miR-307, cgi-miR-745b, cgi-miR-1984 and scaffold1144_2255) increased significantly, while 4 miRNAs (cgi-miR-10a, cgi-miR-10b, cgi-miR-182 and scaffold631_909) decreased significantly in both two comparisons. The similar expression mode of the 9 miRNAs demonstrated that bacteria challenge and heat stress had identical induction effect on their expression level. Heat stress could further improve the response of these miRNAs to bacteria challenge in oyster haemocytes and intensify their immunomodulation during the immune status. The target genes of the 9 miRNAs might be implicated in some physiological functions such as redox reaction and primary energy metabolism, which were essential for the immunity and stress response of mollusc [42]. Therefore, the expression alteration of these miRNAs after heat stress would not hamper the immune response of oysters against invasive pathogens, but further promote their adaption to the two environmental insults. However, the inconsistent expression of remaining 33 miRNAs in the two comparisons indicated that that heat stress could reverse the response of these miRNAs to bacteria challenge in oyster haemocytes and change accordingly their immunomodulation during the immune status. The reversed expression of the 33 miRNAs after heat stress might be detrimental to the immune response in oyster, because heat stress response could compete materials and energy with the immune response for host survival [43]–[45]. The results indicated that heat stress could modulate the immune response through the bilateral induction of immune-related miRNAs to keep maintain homeostasis and increase the adaption.
Materials and Methods
Ethics Statement
The oysters C. gigas (averaging 110 mm in shell height) used in the present study were marine cultured animals, and were collected from a local farm in Qingdao, Shandong Province, China, and maintained in the aerated seawater at 20°C for two weeks before processing. No specific permits are required for the described field studies, since the oysters in the local farm are provided for the local market-sellings. And the oyster C. gigas is not endangered or protected species. All the experiments were conducted according to the regulations of local and central government, and the study protocol was approved by the Experimental Animal Ethics Committee, Institute of Oceanology, Chinese Academy of Sciences, China.
Bacteria Challenge and Heat Stress
One hundred and twenty oysters were employed for the bacteria challenge and heat stress experiment, and they were divided randomly into the control, bacteria and bacteria+heat groups. The oysters in the control and bacteria group received an injection of 100 µL phosphate buffered saline (PBS, 0.14 mol L−1 sodium chloride, 3 mmol L−1 potassium chloride, 8 mmol L−1 disodium hydrogenphosphatedodecahydrate, 1.5 mmol L−1 potassium phosphate monobasic, pH 7.4) and 100 µL suspension of live Vibrio splendidus strain (1×107 CFU ml−1 in PBS), respectively, and maintained in the seawater at 20°C. The oysters in the bacteria+heat group were transferred to 28°C seawater after receiving an injection of 100 µL suspension of live V. splendidus strain (1×107 CFU ml−1, in PBS). All individuals were sampled at 12 h post-injection, and the haemolymph collected from ten individuals were pooled into one sample in each group with three replicates (N = 3) for deep sequencing of small RNA libraries. Meanwhile, five individuals in each group were sampled randomly to collect the haemolymph, and there were five biological replicates (N = 5) for subsequent quantitative real-time PCR analysis of differentially expressed miRNAs. Haemocytes were harvested after the centrifugation at 800× g, 4°C for 10 min, and then resuspended and stored in liquid nitrogen immediately for RNA extraction.
The Construction and Deep Sequencing of Small RNA Libraries
The low molecular weight RNA from the haemocytes of scallop in the control, bacteria and bacteria+heat groups was extracted using a mirVana™ miRNA isolation kit (Ambion, Austin, TX, USA) followed the manufacturer’s protocol. The total RNA in each haemocyte samples was quantified by Nanodrop 2000 (Thermo Scientific) at 260/280 nm (ratio >2.0) and its integrity was checked with Angilent 2100 Bioanalyzer (Agilent Technologies). For the library preparation of each group, the total RNA from 3 samples was mixed equivalently into a RNA sample (∼200 µg) for three technique repeats, and then it was size-fractionated on a 15% tris-borate-EDTA-Urea polyacrylamide gel. After the RNA fragments of 20–30 nucleotides were isolated, the Illumina proprietary adapters were ligated to their 5′ and 3′ terminals, following by reverse transcription. The three generated small cDNA libraries were amplified by PCR with primers complementary to the adaptor sequences. Subsequently, the libraries were deep sequenced by Illumina Hiseq2000 according to the manufacturer’s instructions (Genergy biotechnology company, ShangHai, China). The raw sequencing reads have been submitted to NCBI Short Read Archive under the accession number of SRR1066790–1066792.
The Identification of Oyster miRNAs
Raw reads obtained from Illumina sequencing were processed by the Fastx-toolkit pipeline (http://hannonlab.cshl.edu/fastx_toolkit/index.html) to summarize data production, evaluate sequencing quality, remove low quality reads and adaptor sequences, and calculate the length distribution of small RNA reads [46], [47]. Small RNAs ranging from 18–30 nt in the small RNA library were collected for the removal of redundancy. The resulting clean reads mapped to the RepBase (www.girinst.org/repbase) and Rfam (http://rfam.sanger.ac.uk) were removed before further analysis. And the clean reads mapped to protein-coding mRNA sequences in oyster C. gigas were considered to be degradation products, and therefore, eliminated [31].
Oyster genomic sequences and annotation files are available and downloaded from the Comprehensive Library for Modern Biotechnology (CLiMB) repository (doi:10.5524/100030). After the filtered clean reads were mapped to mature miRNA in miRBase (version 19), the mappable reads were aligned to the oyster genome using bowtie software (http://bowtie-bio.sourceforge.net/index.shtml), and the aligned reads were further analyzed by miRDeep2 software (http://www.mdc-berlin.de/en/research/research_teams/systems_biology_of_gene_regulatory_elements/projects/miRDeep/) with the prediction of hairpin structure and precursor sequence to identify the known miRNAs in oyster. Meanwhile, the unmappable reads with registered mature miRNAs were also analyzed by miRDeep2 software to identify potential novel miRNAs in oyster. Multiple alignment of precursor sequences was performed with the ClustalW multiple alignment program (http://www.ebi.ac.uk/clustalw/).
Expression Analysis of Oyster miRNAs
The copy numbers of known and novel miRNAs in three small RNA libraries were counted using a home-made Perl script. To determine the significant difference of miRNA copy numbers among the three small RNA libraries, the IDEG6 program (http://telethon.bio.unipd.it/bioinfo/IDEG6_form/) was utilized to perform a normalization calculation. Results of the Audice-Claverie test, Fisher exact test and Chi-squared 2×2 test with a Bonferroni correction for multiple comparisons, and a p-value <0.00001 indicated that differences in the miRNA copy numbers were statistically significant [48]. The expression levels of oyster miRNAs in each library were estimated using FPKM method, and a cluster dendrogram of oyster miRNAs with significant expression difference was constructed based on their expression levels using cluster 3.0 software and Treeview software.
Quantitative Real-time PCR Analysis of Differentially Expressed miRNAs
Total RNA including miRNAs in oyster haemocytes were extracted and purified using miRNeasy Mini Kit (QIAGEN) according to its protocol. The reverse transcription of miRNAs was carried out based on QIAGEN miScript II RT Usage information using total RNA as template. The synthesis reaction was performed at 37°C for 1 h, terminated by heating at 95°C for 5 min. The cDNA mix was diluted with the addition of 200 µL RNase-free water for subsequent SYBR Green fluorescent quantitative real-time PCR.
The quantitative real-time PCR was carried out in a total volume of 25.0 µL, containing 12.5 µL of 2x QuantiTect SYBR Green PCR Master Mix (QIAGEN, miScript SYBR Green PCR Kit), 2.5 µL of diluted cDNA, 2.5 µL of each primers (10 mmol L−1), and 5.0 µL of RNase-free water. Eight miRNA fragments were amplified using specific forward primers (Table 3) and universal reverse primers, and the universal reverse primers and 5S primer (Table 3) were used to amplify 5S fragment as an internal control to verify the successful reverse transcription and calibrate the DNA template. The SYBR Green real-time PCR assay was carried out for each sample with three technical replicates in an ABI PRISM 7500 Sequence Detection System (Applied Biosystems). All data was given in terms of relative miRNA expression using the 2−△△Ct method. The data was subjected to one-way analysis of variance (one-way ANOVA) followed by a multiple comparison. Differences were considered significant at P<0.05.
Table 3. Sequence of the primers used in the experiment.
| Primer | Sequence (5′-3′) | Sequence information |
| P1 | TAATACTGTCAGGTAAAGATGTC | Real-time cgi-miR-8 primer |
| P2 | TGAGTATTACATCAGGTACTGA | Real-time cgi-miR-12 primer |
| P3 | AACCCGTAGATCCGAACTTGTG | Real-time cgi-miR-100 primer |
| P4 | TCCCTGAGACCATAACTTGTGA | Real-time cgi-miR-125 primer |
| P5 | TGCCCTATCCGTCAGTCGCTGC | Real-time cgi-miR-1984 primer |
| P6 | AGCTATAATGGTTGTCATTTGTA | Real-time scaffold631_909 primer |
| P7 | CTTGGCACTGTCTGAGCGCAGGT | Real-time scaffold42648_5080 primer |
| P8 | TTGAATTTATTGGTTGGGTGT | Real-time scaffold1599_5643 primer |
| P9 | CAAGGATGACACGCAAAT | Real-time 5S primer |
Target Prediction and GO Analysis
The 3′UTR sequences of oyster protein-coding genes were retrieved based on the oyster genomic sequences and annotation information. The target genes of oyster miRNAs were predicted using the miRanda algorithm (score > = 160, free energy< = −25 kcal/mol, http://www.microrna.org/microrna/getDownloads.do) [49].
For GO analysis, all protein sequences of oyster were aligned by local blastp search to the non-redundant database of NCBI with E-value <1E-5, and then the alignment results were parsed by Blast2GO software (http://www.blast2go.com/b2ghome) for assigning GO terms. The GO terms of genes targeted by the differentially expressed miRNAs were calculated and exhibited through Web Gene Ontology Annotation Plot (WEGO, http://wego.genomics.org.cn/cgi-bin/wego/index.pl).
Supporting Information
The sequence and position information of known miRNAs in oyster.
(XLSX)
The sequence and position information of novel miRNAs in oyster.
(XLSX)
The reads numbers and expression levels of oyster miRNAs in control, bacteria and bacteria+heat libraries.
(XLSX)
The information of differentially expressed miRNAs in the control/bacteria and bacteria/bacteria+heat comparisons.
(XLSX)
All oyster miRNAs and their predicted target genes.
(XLSX)
Acknowledgments
The authors were grateful to all the laboratory members for continuous technical advice and helpful discussion.
Funding Statement
This research was supported by 973 National Key Fundamental Research Program (No. 2010CB126404), National High Technology Research and Development Program (863 Program, No. 2012AA10A401) from the Chinese Ministry of Science and Technology (http://www.most.gov.cn/), grants from NSFC (No. 31072192 to L.W, 30925028 to L.S, http://www.nsfc. gov.cn/), and Shandong Provincial Natural Science Foundation (No. JQ201110 to L.W, http://www.sdnsf.gov.cn/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bushati N, Cohen SM (2007) microRNA functions. Annu Rev Cell Dev Biol 23: 175–205. [DOI] [PubMed] [Google Scholar]
- 2. Davis-Dusenbery BN, Hata A (2010) Mechanisms of control of microRNA biogenesis. J Biochem 148: 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Winter J, Jung S, Keller S, Gregory RI, Diederichs S (2009) Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11: 228–234. [DOI] [PubMed] [Google Scholar]
- 4. Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 5. Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 6. Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Williams AE (2008) Functional aspects of animal microRNAs. Cell Mol Life Sci 65: 545–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leung AK, Sharp PA (2010) MicroRNA functions in stress responses. Mol Cell 40: 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Asgari S (2011) Role of MicroRNAs in Insect Host-Microorganism Interactions. Front Physiol 2: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Y, Li YK (2013) MicroRNAs in the regulation of immune response against infections. Journal of Zhejiang University-Science B 14: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thomas MP, Lieberman J (2013) Live or let die: posttranscriptional gene regulation in cell stress and cell death. Immunol Rev 253: 237–252. [DOI] [PubMed] [Google Scholar]
- 12. Ebert MS, Sharp PA (2012) Roles for microRNAs in conferring robustness to biological processes. Cell 149: 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Christodoulou F, Raible F, Tomer R, Simakov O, Trachana K, et al. (2010) Ancient animal microRNAs and the evolution of tissue identity. Nature 463: 1084–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fullaondo A, Lee SY (2012) Identification of putative miRNA involved in Drosophila melanogaster immune response. Dev Comp Immunol 36: 267–273. [DOI] [PubMed] [Google Scholar]
- 15. O’Neill LA, Sheedy FJ, McCoy CE (2011) MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol 11: 163–175. [DOI] [PubMed] [Google Scholar]
- 16. Ma X, Becker Buscaglia LE, Barker JR, Li Y (2011) MicroRNAs in NF-kappaB signaling. J Mol Cell Biol 3: 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Etebari K, Asgari S (2013) Conserved microRNA miR-8 blocks activation of the Toll pathway by upregulating Serpin 27 transcripts. RNA Biol 10: 1356–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Choi IK, Hyun S (2012) Conserved microRNA miR-8 in fat body regulates innate immune homeostasis in Drosophila. Dev Comp Immunol 37: 50–54. [DOI] [PubMed] [Google Scholar]
- 19. Vaz C, Mer AS, Bhattacharya A, Ramaswamy R (2011) MicroRNAs modulate the dynamics of the NF-kappaB signaling pathway. PLoS One 6: e27774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pocock R (2011) Invited review: decoding the microRNA response to hypoxia. Pflugers Arch 461: 307–315. [DOI] [PubMed] [Google Scholar]
- 21. Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W (2006) Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol 71: 513–521. [DOI] [PubMed] [Google Scholar]
- 22. Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, et al. (2009) The deep evolution of metazoan microRNAs. Evol Dev 11: 50–68. [DOI] [PubMed] [Google Scholar]
- 23. Zhang L, Li L, Zhang G (2011) A Crassostrea gigas Toll-like receptor and comparative analysis of TLR pathway in invertebrates. Fish Shellfish Immunol 30: 653–660. [DOI] [PubMed] [Google Scholar]
- 24. Huan P, Wang H, Dong B, Liu B (2012) Identification of differentially expressed proteins involved in the early larval development of the Pacific oyster Crassostrea gigas. J Proteomics 75: 3855–3865. [DOI] [PubMed] [Google Scholar]
- 25. Zhang G, Fang X, Guo X, Li L, Luo R, et al. (2012) The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490: 49–54. [DOI] [PubMed] [Google Scholar]
- 26. Izar B, Mannala GK, Mraheil MA, Chakraborty T, Hain T (2012) microRNA Response to Listeria monocytogenes Infection in Epithelial Cells. Int J Mol Sci 13: 1173–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. O’Connell RM, Rao DS, Baltimore D (2012) microRNA Regulation of Inflammatory Responses. Annu Rev Immunol 30: 295–312. [DOI] [PubMed] [Google Scholar]
- 28. Li C, Feng W, Qiu L, Xia C, Su X, et al. (2012) Characterization of skin ulceration syndrome associated microRNAs in sea cucumber Apostichopus japonicus by deep sequencing. Fish Shellfish Immunol 33: 436–441. [DOI] [PubMed] [Google Scholar]
- 29. Cristino AS, Tanaka ED, Rubio M, Piulachs MD, Belles X (2011) Deep sequencing of organ- and stage-specific microRNAs in the evolutionarily basal insect Blattella germanica (L.) (Dictyoptera, Blattellidae). PLoS One 6: e19350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wei Y, Chen S, Yang P, Ma Z, Kang L (2009) Characterization and comparative profiling of the small RNA transcriptomes in two phases of locust. Genome Biol 10: R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Etebari K, Hussain M, Asgari S (2013) Identification of microRNAs from Plutella xylostella larvae associated with parasitization by Diadegma semiclausum. Insect Biochem Mol Biol 43: 309–318. [DOI] [PubMed] [Google Scholar]
- 32. Berezikov E (2011) Evolution of microRNA diversity and regulation in animals. Nat Rev Genet 12: 846–860. [DOI] [PubMed] [Google Scholar]
- 33. Starega-Roslan J, Koscianska E, Kozlowski P, Krzyzosiak WJ (2011) The role of the precursor structure in the biogenesis of microRNA. Cell Mol Life Sci 68: 2859–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xiao C, Rajewsky K (2009) MicroRNA control in the immune system: basic principles. Cell 136: 26–36. [DOI] [PubMed] [Google Scholar]
- 35. Schmidt WM, Spiel AO, Jilma B, Wolzt M, Muller M (2009) In vivo profile of the human leukocyte microRNA response to endotoxemia. Biochem Biophys Res Commun 380: 437–441. [DOI] [PubMed] [Google Scholar]
- 36. Chen YH (2011) MicroRNA immunobiology: when microRNA chemists meet immunologists. Cell Mol Immunol 8: 369–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bronevetsky Y, Ansel KM (2013) Regulation of miRNA biogenesis and turnover in the immune system. Immunol Rev 253: 304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McCall CE, El Gazzar M, Liu T, Vachharajani V, Yoza B (2011) Epigenetics, bioenergetics, and microRNA coordinate gene-specific reprogramming during acute systemic inflammation. J Leukoc Biol 90: 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weitzel RP, Lesniewski ML, Haviernik P, Kadereit S, Leahy P, et al. (2009) microRNA 184 regulates expression of NFAT1 in umbilical cord blood CD4+ T cells. Blood 113: 6648–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen Y, Stallings RL (2007) Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res 67: 976–983. [DOI] [PubMed] [Google Scholar]
- 41. Yu X, Wang H, Lu Y, de Ruiter M, Cariaso M, et al. (2012) Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J Exp Bot 63: 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang X, Wang L, Zhang H, Ji Q, Song L, et al. (2012) Immune response and energy metabolism of Chlamys farreri under Vibrio anguillarum challenge and high temperature exposure. Fish Shellfish Immunol 33: 1016–1026. [DOI] [PubMed] [Google Scholar]
- 43. Malham SK, Lacoste A, Gelebart F, Cueff A, Poulet SA (2003) Evidence for a direct link between stress and immunity in the mollusc Haliotis tuberculata. Journal of Experimental Zoology Part a-Comparative Experimental Biology 295A: 136–144. [DOI] [PubMed] [Google Scholar]
- 44. Lacoste A, Malham SK, Cueff A, Poulet SA (2001) Noradrenaline modulates hemocyte reactive oxygen species production via beta-adrenergic receptors in the oyster Crassostrea gigas. Dev Comp Immunol 25: 285–289. [DOI] [PubMed] [Google Scholar]
- 45. Lacoste A, Jalabert F, Malham SK, Cueff A, Poulet SA (2001) Stress and stress-induced neuroendocrine changes increase the susceptibility of juvenile oysters (Crassostrea gigas) to Vibrio splendidus. Appl Environ Microbiol 67: 2304–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xu Z, Chen J, Li X, Ge J, Pan J, et al. (2013) Identification and characterization of microRNAs in channel catfish (Ictalurus punctatus) by using Solexa sequencing technology. PLoS One 8: e54174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Song QX, Liu YF, Hu XY, Zhang WK, Ma B, et al. (2011) Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol 11: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ou J, Meng Q, Li Y, Xiu Y, Du J, et al. (2012) Identification and comparative analysis of the Eriocheir sinensis microRNA transcriptome response to Spiroplasma eriocheiris infection using a deep sequencing approach. Fish Shellfish Immunol 32: 345–352. [DOI] [PubMed] [Google Scholar]
- 49. Milagro FI, Miranda J, Portillo MP, Fernandez-Quintela A, Campion J, et al. (2013) High-throughput sequencing of microRNAs in peripheral blood mononuclear cells: identification of potential weight loss biomarkers. PLoS One 8: e54319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sequence and position information of known miRNAs in oyster.
(XLSX)
The sequence and position information of novel miRNAs in oyster.
(XLSX)
The reads numbers and expression levels of oyster miRNAs in control, bacteria and bacteria+heat libraries.
(XLSX)
The information of differentially expressed miRNAs in the control/bacteria and bacteria/bacteria+heat comparisons.
(XLSX)
All oyster miRNAs and their predicted target genes.
(XLSX)