Abstract
Background
Histoplasma capsulatum and Pneumocystis organisms cause host infections primarily affecting the lung tissue. H. capsulatum is endemic in the United States of America and Latin American countries. In special environments, H. capsulatum is commonly associated with bat and bird droppings. Pneumocystis-host specificity has been primarily studied in laboratory animals, and its ability to be harboured by wild animals remains as an important issue for understanding the spread of this pathogen in nature. Bats infected with H. capsulatum or Pneumocystis spp. have been found, with this mammal serving as a probable reservoir and disperser; however, the co-infection of bats with both of these microorganisms has never been explored. To evaluate the impact of H. capsulatum and Pneumocystis spp. infections in this flying mammal, 21 bat lungs from Argentina (AR), 13 from French Guyana (FG), and 88 from Mexico (MX) were screened using nested-PCR of the fragments, employing the Hcp100 locus for H. capsulatum and the mtLSUrRNA and mtSSUrRNA loci for Pneumocystis organisms.
Results
Of the 122 bats studied, 98 revealed H. capsulatum infections in which 55 of these bats exhibited this infection alone. In addition, 51 bats revealed Pneumocystis spp. infection of which eight bats exhibited a Pneumocystis infection alone. A total of 43 bats (eight from AR, one from FG, and 34 from MX) were found co-infected with both fungi, representing a co-infection rate of 35.2% (95% CI = 26.8-43.6%).
Conclusion
The data highlights the H. capsulatum and Pneumocystis spp.co-infection in bat population’s suggesting interplay with this wild host.
Keywords: Histoplasma, Pneumocystis, Co-infection, Bats, PCR
Background
Histoplasmosis due to Histoplasma capsulatum and pneumonia caused by Pneumocystis spp. are fungal diseases that can generate serious and even life-threatening pneumonia in immunosuppressed hosts. H. capsulatum is a fungal pathogen that affects a wide range of mammal species, including the human. Autochthonous clinical cases have been reported between the latitudes 54° 05′ North (Alberta, Canada) and 38° South (Neuquén, Argentina) [1,2]. The disease associated with this fungus is relevant in the geographical areas where histoplasmosis is endemic or epidemic, such as the Missouri, Ohio, and Mississippi river valleys, in the United States of America (USA), and some Latin American countries with a high frequency of outbreaks [3,4]. In Mexico, histoplasmosis is widely distributed and case reports are rather variable [4]. Infection is caused by the inhalation of fungal saprobe mycelial-phase propagules (infective form) that develop in special environments and are mainly found in bat guano accumulated in confined spaces such as caves and abandoned mines and buildings. The potential role of bats in spreading H. capsulatum in nature remains unclear. The high risk of natural bat infection with this fungus in Mexican caves has been well-documented [5-8]. According to their genetic diversities, H. capsulatum isolates from different geographical origins have been grouped into eight clades; seven of which are considered phylogenetic species. Among these, highlight the LAm A clade that harbours significant genetic variability [9].
The genus Pneumocystis contains highly diversified fungal pathogens that are harboured by a wide range of mammal hosts [10-16]. Pneumocystis organisms, which are transmitted via host-to-host airborne route, have a marked host-species-related diversity that is associated with close host specificity. The high divergence among Pneumocystis species most likely resulted from a prolonged process of co-evolution with each mammal host, mostly associated with co-speciation, as suggested by Demanche et al. [12] and Hugot et al. [13]. Although most phenotypic and genotypic data supporting Pneumocystis stenoxenism derives from laboratory animal models or captive animals, reports about Pneumocystis prevalence and circulation in wild fauna are scarce [12-16].
Unpublished preliminary data by our team revealed H. capsulatum and Pneumocystis co-infection in two randomly captured bats, identifying these mammals as probable reservoirs and dispersers of both parasites in nature (Dei-Cas E and Taylor ML, comm. pers.). The study of co-infection systems, where the host (i.e. a wild host) usually harbours two or multiple parasites, requires an in-depth investigation to determine a comprehensive understanding of this multi-infectious process in regards to its dynamics and consequences.
H. capsulatum and Pneumocystis share a number of features that justify their concomitant study, including: a low pathogenicity in healthy hosts and severe disease in immunocompromised hosts; an induced immune response; a respiratory portal of entry; and the ability to disseminate from the lungs to other organs. Therefore, it is possible that co-infection between both parasites is highly common in nature.
The aim of the present research was to detect the frequency of the H. capsulatum and Pneumocystis organisms’ infection and co-infection in the lung samples of a number of wild bat species from three countries from Latin America. For this purpose, we used a highly sensitive PCR with specific molecular markers for each pathogen that have been used successfully in clinical patients.
Methods
Bat samples
A total of 122 bats from different species and families were randomly captured as reported by Taylor et al. [7]: 21 came from Argentina; 13 came from French Guyana; and 88 came from Mexico. In all cases national rules regulating bat species protection, capture, and processing have adhered to strict ethical recommendations and to the guidelines published by Gannon, Sikes and the Animal Care and Use Committee of the American Society of Mammalogists [17].
The bats were euthanized by cervical dislocation and processed according to recommendations and approval of the Faculty of Medicine Ethics Committee, in accordance with the Animal Care and Use Committee of the UNAM and the Mexican Official Guide (NOM 062-ZOO-1999). The lungs from each bat captured in Mexico were separated and immediately frozen at −20°C. Animals captured in Argentina and French Guyana were also euthanized by cervical dislocation and processed in situ and their lungs were separated and preserved in 70% ethanol until DNA extraction.
DNA samples
DNA was extracted from the bat lungs using a DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, USA). After extraction, the DNA samples were frozen at −20°C. The DNA samples were screened for H. capsulatum infection using nested-PCR for a fragment of the gene encoding a 100-kDa protein (Hcp100) [18], a molecular marker considered to be highly specific for this pathogen. Molecular screening for Pneumocystis spp. infection was conducted in parallel, using nested-PCR for fragments of the rRNA mitochondrial large [19,20] and small [21] subunit loci, mtLSUrRNA and mtSSUrRNA, respectively.
Nested PCR assay of the Hcp100 locus for the detection of H. capsulatum
The assay was performed as described by Bialek et al. [18] with minor modifications by Taylor et al. [22] that did not change the specificity and sensitivity of the Hcp100 marker. Two sets of primers, described by Bialek et al. [18], were used: the outer primer set included HcI (5′-GCG-TTC-CGA-GCC-TTC-CAC-CTC-AAC-3′) and HcII (5′-ATG-TCC-CAT-CGG-GCG-CCG-TGT-AGT-3′); the inner primers were HcIII (5′-GAG-ATC-TAG-TCG-CGG-CCA-GGT-TCA-3′) and HcIV (5′-AGG-AGA-GAA-CTG-TAT-CGG-TGG-CTT-G-3′) and delimit a 210 base pair (bp) fragment unique to H. capsulatum. The primers were supplied by Operon Technologies Inc. (Alameda, CA, USA).
The first and second PCR reactions of the Hcp100 locus were standardised elsewhere [6]. For the first round of amplification, the thermocycling conditions were as follows: one cycle at 94°C for 5 min; 35 cycles at 94°C for 30 s, 50°C for 30 s and 72°C for 1 min; and a final cycle at 72°C for 5 min. Thermocycling conditions for the second PCR (nested reaction) were: one cycle at 94°C for 5 min; 30 cycles at 94°C for 30 s, 65°C for 30 s and 72°C for 1 min; and a final extension cycle at 72°C for 5 min. The DNA (20 ng) of the EH-53 H. capsulatum strain from a Mexican clinical case was used as a positive amplification control, and Milli-Q water was processed as a negative control.
Nested PCR assays of the mtLSUrRNA and mtSSUrRNA loci for the detection of Pneumocystis spp
The assays were based on amplifying fragments of the mitochondrial large (mtLSU) and small (mtSSU) subunits of the rRNA gene.
Nested PCR at the mtLSUrRNA locus employed the outer primer set published by Wakefield et al. [19], pAZ102-H (5′-GTG-TAC-GTT-GCA-AAG-TAG-TC-3′) and pAZ102-E (5′-GAT-GGC-TGT-TTC-CAA-GCC-CA-3′). The inner primers pAZ102-X (5′-GTG-AAA-TAC-AAA-TCG-GAC-TAG-G-3′) and pAZ102-Y (5′-TCA-CTT-AAT-ATT-AAT-TGG-GGA-GC-3′) and delimit a 267 bp fragment for Pneumocystis[20]. The first and nested PCR reactions of the mtLSUrRNA locus were standardised as described elsewhere [14,19,20]. For the first round of amplification, the thermocycling conditions were as follows: 30 cycles at 94°C for 30 s, 50°C for 1 min, and 65°C for 1 min. The nested reaction was performed with 10% of the first-round amplification product and the thermocycling conditions were: 30 cycles at 94°C for 30 s, 55°C for 1 min, and 65°C for 1 min.
Nested PCR at the mtSSUrRNA locus was performed with the outer primers, pAZ112-10 F (5′-GGG-AAT-TCT-AGA-CGG-TCA-CAG-AGA-TCA-G-3′) and pAZ112-10R (5′-GGG-AAT-TCG-AAC-GAT-TAC-TAG-CAA-CCC-3′). The inner primers pAZ112-13RI (5′-GGG-AAT-TCG-AAG-CAT-GTT-GTT-TAA-TTC-G-3′) and pAZ112-14RI (5′-GGG-AAT-TCT-TCA-AAG-AAT-CGA-GTT-TCA-G-3′) and delimit a 300 bp fragment for Pneumocystis species, as reported by Tsolaki et al. [21]. PCR for the mtSSUrRNA locus was previously described by Tsolaki et al. [21] and Akbar et al. [14]. For the first round of amplification, the thermocycling conditions were as follows: 40 cycles at 94°C for 30 s, 55°C for 1 min, and 65°C for 1 min. The nested round was performed with 10% of the first-round amplification product and the thermocycling conditions were: 10 cycles at 94°C for 30 s, 52°C for 1 min, and 65°C for 1 min, followed by 30 cycles at 94°C for 30 s, 63°C for 1 min, and 65°C for 1 min.
Primers for the mtLSUrRNA and mtSSUrRNA loci were supplied by Operon Technologies. The DNA (20 ng) of rabbit Pneumocystis (Pneumocystis oryctolagi) was processed as a positive amplification control, and Milli-Q water was used as a negative control for both Pneumocystis molecular markers.
Amplified products
Amplicons from each PCR assay were electrophoresed through 1.5% agarose in 0.5X Tris-borate-EDTA buffer. Electrophoresis was conducted at 120 V for 50 min. The 100 bp DNA Ladder (Gibco Laboratories, Grand Island, NY, USA) was used as a molecular size marker. The bands were visualised using a UV transilluminator after ethidium bromide staining (0.5 μg/mL). The amplicons were purified using the QIAquick® PCR and the QIAEX II kits (Qiagen) for the H. capsulatum and Pneumocystis organisms, respectively. Afterwards, the amplicons were sent to the Molecular Biology Laboratory, Institute of Cellular Physiology (UNAM, Mexico) for sequencing in an ABI-automated DNA sequencer (Applied Biosystems Inc., Foster City, CA, USA). Sequencing reactions were performed for forward and reverse DNA strands, and a consensus sequence for each amplified bat lung sample product was generated. The sequences were edited and aligned using the MEGA software, version 5 (http://www.megasoftware.net).
Most of the Hcp100 sequences of H. capsulatum were previously reported in González-González et al. [6], and the other sequences were deposited in a database [GenBank: from JX091346 to JX091370 accession numbers]. All sequences generated by both molecular markers for Pneumocystis spp. were reported by Derouiche et al. [16] and Akbar et al. [14].
The sequences of the specific markers for each pathogen (i.e., Hcp100 for H. capsulatum and mtLSUrRNA or mtSSUrRNA for Pneumocystis spp.) that were obtained in the same animal were the main inclusion criterion for considering bat co-infection.
Statistics
The infection and co-infection rates for each pathogen were estimated by considering all of the bats studied from the three countries and from each country separately (Argentina, French Guyana, and Mexico), in relation to those bats with H. capsulatum and Pneumocystis spp. infections as identified by sequencing their respective molecular markers. The corresponding 95% confidence interval (CI) was calculated using a normal distribution.
Results
Data from nine bat species studied belonging to five different families, highlighting their particular behaviours, such as migration, nourishment, distribution in the American continent and colony size, are referred to in Table 1, according to Ceballos and Oliva [23]. These behaviours varied considerably among the bat species studied (Table 1). The different species captured, their numbers, and their geographical origins are registered in Table 2. Although most of the bat species studied were non-migratory, the number of migratory bats from three processed species was greater than that of the non-migratory species (Tables 1 and 2). It is noteworthy that among the 122 bats studied, 84 (68.80%) belonged to the migratory species Tadarida brasiliensis, from which 63 individuals were captured in Mexico and 21 in Argentina (Table 2).
Table 1.
Families, species, and behaviours of the bats studied
| (Family) Species | Migration | Nourishment | Distribution | Colony size |
|---|---|---|---|---|
| (Phyllostomidae) |
|
|
|
|
|
Artibeus hirsutus |
Non-migratory |
Frugivorous |
From south of Sonora to south of Guerrero, Mexico |
Not defined |
|
Carollia perspicillata |
Non-migratory |
Frugivorous |
From Tamaulipas to Oaxaca, Mexico, and to south of Bolivia, Brazil and Paraguay |
Small groups from 10 to 100 individuals |
|
Glossophaga soricina |
Non-migratory |
Nectarivorous, polinivorous, frugivorous and insectivorous |
From Mexico to South America |
From a few to 2,000 individuals |
| (Natalidae) |
|
|
|
|
|
Natalus stramineus |
Non-migratory |
Insectivorous |
From north of Mexico to Brazil |
Approximately 10,000 individuals |
| (Mormoopidae) |
|
|
|
|
|
Pteronotus davyi |
Non-migratory |
Insectivorous |
From north of Mexico to Brazil |
From hundreds to thousands of individuals |
|
Pteronotus parnellii |
Non-migratory |
Insectivorous |
From north of Mexico to Brazil |
From hundreds to thousands of individuals |
|
Pteronotus parnellii |
Non-migratory |
Insectivorous |
From north of Mexico to the north of Argentina and Paraguay |
Thousands of individuals |
|
Mormoops megalophylla |
Migratory |
Insectivorous |
From south USA to Venezuela and Peru |
From a few to thousands of individuals |
| (Molossidae) |
|
|
|
|
|
Tadarida brasiliensis |
Migratory |
Insectivorous |
From central USA to Chile and Argentina |
Generally, thousands to millions of individuals |
| (Vespertilionidae) |
|
|
|
|
| Myotis californicus | Occasionally migratory | Insectivorous | From western Canada and USA to Guatemala | Small groups or gregarious |
Data from the bat species were reported by Ceballos and Oliva [23].
Table 2.
Species, numbers, and geographical origins of the bats infected with H. capsulatum or Pneumocystis spp.
|
Species |
Geographical origins/localities |
Number of bats infected with
H. capsulatum
/
Pneumocystis
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| |
Argentina |
French Guyana |
Mexico |
(Total samples per species) |
||||||
| |
TUC |
CBA |
Kourou |
CS |
MN |
GR |
HG |
MS |
NL |
|
| H. capsulatum / Pneumocystis (total samples) | ||||||||||
|
A. hirsutus |
|
|
|
|
|
|
|
5/3 (5) |
|
5/3 (5) |
|
C. perspicillata |
|
|
1/0 (1) |
|
|
|
|
|
|
1/0 (1) |
|
G. soricina |
|
|
3/6 (12) |
|
|
4/3 (4) |
|
|
|
7/9 (16) |
|
N. stramineus |
|
|
|
|
|
5/1 (8) |
|
|
|
5/1 (8) |
|
P. davyi |
|
|
|
|
|
1/0 (1) |
|
|
|
1/0 (1) |
|
P. parnellii |
|
|
|
|
|
2/0 (2) |
0/1 (1) |
|
|
2/1 (3) |
|
M. megalophylla |
|
|
|
|
|
2/0 (2) |
|
|
1/0 (1) |
3/0 (3) |
|
T. brasiliensis |
16/8 (16) |
3/ND (5) |
|
8/2 (8) |
7/2 (8) |
|
13/5 (20) |
|
26/19 (27) |
73/36 (84) |
|
M. californicus |
|
|
|
|
|
|
|
|
1/1 (1) |
1/1 (1) |
| Number of bats infected with H. capsulatum/Pneumocystis (Total samples per locality) | 16/8 (16) | 3/ND (5) | 4/6 (13) | 8/2 (8) | 7/2 (8) | 14/4 (17) | 13/6 (21) | 5/3 (5) | 28/20 (29) | 98/51 (122) |
Abbreviations: TUC = Tucumán; CBA = Córdoba; CS = Chiapas; MN = Michoacán; GR = Guerrero; HG = Hidalgo; MS = Morelos; NL = Nuevo León.
ND = Not determined.
Detection of H. capsulatum infection in the bat lung samples
Of the 122 bat lungs that were molecularly screened for H. capsulatum infection, 98 bats generated sequences for the Hcp100 marker, of which 55 bats were found to be infected with this pathogen alone, corresponding to 45.1% (95% CI = 36.4-53.6%) of the 122 bats from the three geographical regions studied (Figure 1).
Figure 1.
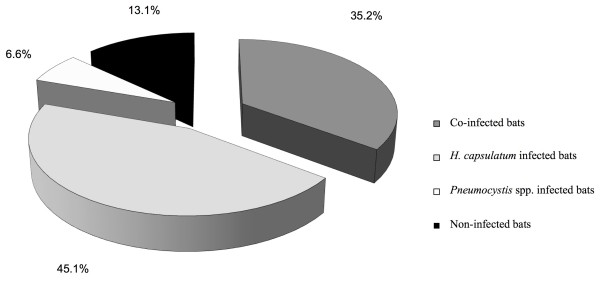
Percentages of H. capsulatum and Pneumocystis infection and their respective co-infection in bats randomly sampled in Argentina, French Guyana, and Mexico. Each percentage value was calculated based on the 122 bats captured. Bat infection was screened with specific molecular markers for each pathogen, as described in the Methods section.
Table 2 displays the number of infected bats with H. capsulatum in relation to the total number of each bat species sampled at different localities from the monitored Latin American countries.
Detection of Pneumocystis spp. infection in the bat lung samples
Of the 122 lungs that were molecularly screened for Pneumocystis spp., 51 bats generated sequences for one or both of the Pneumocystis molecular markers assayed. From these sequences, seven matched the mtLSUrRNA locus and another seven matched the mtSSUrRNA locus, while 37 sequences were generated at both loci. Pneumocystis spp. infection alone was found only in eight bats, corresponding to 6.6% (95% CI = 2.25-10.85%) of the total bats studied (Figure 1).
Table 2 displays the number of infected bats with Pneumocystis spp. in relation to the total number of each bat species sampled at different localities from the monitored Latin American countries.
H. capsulatum and Pneumocystis spp. co-infection in the bat lung samples
Of the lung samples from the 122 bats captured in Argentina, French Guyana, and Mexico that were molecularly screened for H. capsulatum and Pneumocystis infections, 43 samples revealed the specific sequences of each pathogen, corresponding to 35.2% (95% CI = 26.8-43.6%) of the samples being co-infected with both pathogens in bats from the three geographical regions studied (Figure 1).
Table 3 displays the number of co-infected bats with both pathogens in relation to the total number of each bat species sampled at different localities from the monitored Latin American countries.
Table 3.
Species, numbers, and geographical origins of the bats co-infected with H. capsulatum and Pneumocystis spp.
|
Species |
Geographical origins/localities |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| |
Argentina (n = 21) |
French Guyana (n = 13) |
Mexico (n = 88) |
Number of co-infected bats |
||||||
| |
TUC |
CBA |
Kourou |
CS |
MN |
GR |
HG |
MS |
NL |
(Total samples per species) |
| Co-infection (total samples) | ||||||||||
|
A. hirsutus |
|
|
|
|
|
|
|
3 (5) |
|
3 (5) |
|
C. perspicillata |
|
|
0 (1) |
|
|
|
|
|
|
0 (1) |
|
G. soricina |
|
|
1 (12) |
|
|
3 (4) |
|
|
|
4 (16) |
|
N. stramineus |
|
|
|
|
|
1 (8) |
|
|
|
1 (8) |
|
P. davyi |
|
|
|
|
|
0 (1) |
|
|
|
0 (1) |
|
P. parnellii |
|
|
|
|
|
0 (2) |
0 (1) |
|
|
0 (3) |
|
M. megalophylla |
|
|
|
|
|
0 (2) |
|
|
0 (1) |
0 (3) |
|
T. brasiliensis |
8 (16) |
0 (5) |
|
2 (8) |
2 (8) |
|
3 (20) |
|
19 (27) |
34 (84) |
|
M. californicus |
|
|
|
|
|
|
|
|
1 (1) |
1 (1) |
| Number of co-infected bats (Total samples per locality) | 8 (16) | 0 (5) | 1 (13) | 2 (8) | 2 (8) | 4 (17) | 3 (21) | 3 (5) | 20 (29) | 43 (122) |
Abbreviations: TUC = Tucumán; CBA = Córdoba; CS = Chiapas; MN = Michoacán; GR = Guerrero; HG = Hidalgo; MS = Morelos; NL = Nuevo León.
Finally, of the total number of bat lungs sampled, 106 (86.8%, 95% CI = 80.92-92.68%) were found to be infected with one or both pathogens, whereas 16 (13.1%, 95% CI = 7.22-18.98%) did not show evidence of infection with any of the pathogens studied (Figure 1).
Discussion
The co-infection relationship between the host and different parasite species could occur in natural conditions, although it has been scarcely studied due to its complexity and poor understanding [24]. The presence of more than one parasite species in a single host can lead to positive or negative interactions. In the positive interaction, the parasite could favour the entry and survival of another parasite, whereas in the negative interaction the establishment of a parasite prevents the entry of other parasites and abolishes their survival [24]. It is well accepted in medical research that the infection concept implies the presence of the pathogen in the infected host’s tissues, which does not necessarily indicate a disease status that is supported by characteristic signs and symptoms. Although bats, in general, have a high infection rate with H. capsulatum in their shelters, they most likely do not develop a severe course of the disease [7], and the impact of this infection on the survival of their population is unknown. With regard to Pneumocystis bat infection, this wild host could present a latent infection without evidence of any disease signs and symptoms [12,14]. Consequently, bats could be potential carriers of both parasites in the environment.
H. capsulatum and Pneumocystis spp. cause a host infection through the respiratory airway, mainly affecting the pulmonary tissue. After infecting the lungs, each parasite develops on distinct host environments and exploits different host resources. The H. capsulatum parasitic yeast-phase is an intracellular pathogen of the lung phagocytic cells. In contrast, Pneumocystis organisms are extracellular pathogens that frequently attach to type I pneumocytes [10].
Histoplasma-Pneumocystis co-infection has been reported in immunosuppressed human patients [25], whereas reports of co-infection in wild mammals have not been published. This fact should be re-examined because both parasites are able to share the same wild hosts in a particular manner, likely associated with the host immune status related to stress, sickness, and nutrient starvation.
PCR assays that utilize specific molecular markers are very sensitive tools for detecting a low fungal burden in clinical samples from asymptomatic patients. Currently, H. capsulatum and Pneumocystis spp. infections are detected by different PCR methods, either in human clinical cases or in experimental models [14,26-29]. The present study is the first report for detecting a natural co-infection in wild bats from three distant geographical Latin American regions, using specific PCR assay for each parasite.
The numbers of wild bats infected with H. capsulatum or Pneumocystis organisms varied, with the number of H. capsulatum infected bats surpassing the number of Pneumocystis infected bats (Figure 1). No association was found between a bat species’ susceptibility and nourishment and the rate of infection with both pathogens. However, it is possible that some bat behaviours promote different infection rates for either H. capsulatum or Pneumocystis spp. According to published findings, the rates of each pathogen infection could be associated with the bat colony size and their movements, in the case of H. capsulatum[7], or with behavioural factors such as bats crowding and migration in the case of Pneumocystis spp. [14]. The biggest colonies, mainly of T. brasiliensis, had the highest rate of infection with H. capsulatum, most likely due to bat colony movements within enclosed spaces, especially in shelters where short ceiling-to-floor distances prevails, which facilitate the development of a great number of airborne infective propagules on the abundant guano accumulated underneath bat colonies [7]. Hence, each of these factors allows the co-infection state with both pathogens.
Based on the following evidence, it is likely that either H. capsulatum or Pneumocystis displayed an interaction with different bat species since million of years ago (Ma): 1.- Bat fossils (Tadarida sp.) were reported approximately 3.6 – 1.8 Ma in the Late Pliocene [30]; 2.- the H. capsulatum complex most likely started its radiation at 13–3 Ma in the Miocene [9]; and 3.- the Pneumocystis species have had interaction with mammal hosts for more than 100 Ma [10-13,31]. Under this assumption, the co-infection of both pathogens most likely generated a co-evolution process between each pathogen and the wild host.
Data pertaining to Histoplasma-Pneumocystis co-infection reveal a rate of 35.2%; this finding could be useful for understanding the persistence of both infections in susceptible hosts. The absence of Histoplasma or Pneumocystis infections in 13.1% of the bats studied could suggest that most of the analysed bat populations were exposed to a high risk of infection with these pathogens in their shelters. Co-infection interactions could cause ecological and immunological implications for the host. For the ecological implications, space and alimentary competitions are involved. For the immunological implications, the host immune response against H. capsulatum at the pulmonary level involves cells and molecules that could also participate in the host immune response against Pneumocystis, or vice versa.
Conclusion
The impact of the present findings highlights the H. capsulatum and Pneumocystis spp. co-infection in bat population’s suggesting interplay with this wild host. In addition, this co-infection state could also interfere with the outcome of the disease associated with each pathogen.
Competing interests
The authors declare that they have no conflicts of interest.
Authors’ contributions
MLT and EDC contributed equally to the design of this study. AEGG coordinated and performed the molecular assays for H. capsulatum detection. MLT and AEGG contributed equally to draft the manuscript. JARB and LECB processed the bat samples from Argentina and Mexico and collaborated in the molecular assays for H. capsulatum. EDC, ELMA, CMAD, CD, and MC coordinated the molecular assays of Pneumocystis and revised the manuscript draft. MP, HA, and SD performed molecular assays for Pneumocystis detection. All authors have read and approved the manuscript.
Contributor Information
Antonio E González-González, Email: anto.egg@gmail.com.
Cécile M Aliouat-Denis, Email: cecile.aliouat@univ-lille2.fr.
José A Ramírez-Bárcenas, Email: ton1005an@netscap.net.
Christine Demanche, Email: christine.demanche@univ-lille2.fr.
Muriel Pottier, Email: muriel.pottier@univ-lille2.fr.
Laura E Carreto-Binaghi, Email: lauraelena_c@yahoo.com.
Haroon Akbar, Email: drharoonakbar@yahoo.co.uk.
Sandra Derouiche, Email: sandra.derouiche@etu.univ-lille2.fr.
Magalie Chabé, Email: magali.chabe@univ-lille2.fr.
El Moukhtar Aliouat, Email: elmoukhtar.aliouat-3@univ-lille2.fr.
Eduardo Dei-Cas, Email: eduardo.dei-cas@pasteur-lille.fr.
Maria Lucia Taylor, Email: emello@unam.mx.
Acknowledgements
Dr. M. L. Taylor thanks L. J. López and A. Gómez Nísino from the Instituto de Ecología, UNAM, for their help with accessing several Mexican caves, with bat captures and taxonomic determination, and acknowledges the extraordinary help of Dr. R. Bárquez from the Instituto Lillo to access the Dique Escaba, San Miguel de Tucumán, Tucumán, Argentina. The authors thank I. Mascher for editorial assistance. The present research was supported by the following grants: SEP-CONACYT-ANUIES-ECOS-M05-A03 (Mexico-France) and DGAPA-PAPIIT -Ref. No. IN-203407-3, UNAM, Mexico. A. E. González-González thanks the Biological Science Graduate Program of UNAM and the scholarship of CONACYT (Ref. No. 23492).
References
- Anderson H, Honish L, Taylor G, Johnson M, Tovstiuk C, Fanning A, Tyrrell G, Rennie R, Jaipaul J, Sand C, Probert S. Histoplasmosis cluster, golf course, Canada. Emerg Infect Dis. 2006;12:163–165. doi: 10.3201/eid1201.051083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calanni LM, Pérez R, Brasili S, Schmidt NG, Iovannitti CA, Zuiani MF, Negroni R, Finquelievich J, Canteros CE. Brote de histoplasmosis en la Provincia de Neuquén, Patagonia Argentina. Rev Iberoam Micol. 2013. doi:10.1016/j.riam.2012.12.007. [DOI] [PubMed]
- Guimarães AJ, de Cerqueira MD, Nosanchuk JD. Surface architecture of Histoplasma capsulatum. Front Microbiol. 2011;2:225. doi: 10.3389/fmicb.2011.00225. doi: 10.3389/fmicb.2011.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor ML, Reyes-Montes MR, Chávez-Tapia CB, Curiel-Quesada E, Duarte-Escalante E, Rodríguez-Arellanes G, Peña-Sandoval GR, Valenzuela-Tovar F. In: Research Advances in Microbiology. Benedik M, editor. Kerala: Global Research Network; 2000. Ecology and molecular epidemiology findings of Histoplasma capsulatum, in Mexico; pp. 29–35. [Google Scholar]
- Chávez-Tapia CB, Vargas-Yáñez R, Rodríguez-Arellanes G, Peña-Sandoval GR, Flores-Estrada JJ, Reyes-Montes MR, Taylor ML. I. El murciélago como reservorio y responsable de la dispersión de Histoplasma capsulatum en la naturaleza. II. Papel de los marcadores moleculares del hongo aislado de murciélagos infectados. Rev Inst Nal Enf Resp Mex. 1998;11:187–191. [Google Scholar]
- González-González AE, Aliouat-Denis CM, Carreto-Binaghi LE, Ramírez JA, Rodríguez-Arellanes G, Demanche C, Chabé M, Aliouat EM, Dei-Cas E, Taylor ML. An Hcp100 gene fragment reveals Histoplasma capsulatum presence in lungs of Tadarida brasiliensis migratory bats. Epidemiol Infect. 2012;140:1955–1963. doi: 10.1017/S0950268811002585. [DOI] [PubMed] [Google Scholar]
- Taylor ML, Chávez-Tapia CB, Vargas-Yáñez R, Rodríguez-Arellanes G, Peña-Sandoval GR, Toriello C, Pérez A, Reyes-Montes MR. Environmental conditions favoring bat infections with Histoplasma capsulatum in Mexican shelters. Am J Trop Med Hyg. 1999;61:914–919. doi: 10.4269/ajtmh.1999.61.914. [DOI] [PubMed] [Google Scholar]
- Taylor ML, Hernández-García L, Estrada-Bárcenas D, Salas-Lizana R, Zancopé-Oliveira RM, García De La Cruz S, Galvao-Dias MA, Curiel-Quesada E, Canteros CE, Bojórquez-Torres G, Bogard-Fuentes CA, Zamora-Tehozol E. Genetic diversity of Histoplasma capsulatum isolated from infected bats randomly captured in Mexico, Brazil, and Argentina, using the polymorphism of (GA)n microsatellite and its flanking regions. Fungal Biol. 2012;116:308–317. doi: 10.1016/j.funbio.2011.12.004. [DOI] [PubMed] [Google Scholar]
- Kasuga T, White TJ, Koenig G, McEwen J, Restrepo A, Castañeda E, Da Silva-Lacaz C, Heins-Vaccari EM, De Freitas RS, Zancopé-Oliveira RM, Zhenyu Q, Negroni R, Carter DA, Mikami Y, Tamura M, Taylor ML, Miller GF, Poonwan N, Taylor JW. Phylogeography of the fungal pathogen Histoplasma capsulatum. Mol Ecol. 2003;12:3383–3401. doi: 10.1046/j.1365-294x.2003.01995.x. [DOI] [PubMed] [Google Scholar]
- Aliouat-Denis CM, Chabé M, Demanche C, Aliouat EM, Viscogliosi E, Guillot J, Delhaes L, Dei-Cas E. Pneumocystis species, co-evolution and pathogenic power. Infect Genetic Evol. 2008;8:708–726. doi: 10.1016/j.meegid.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Guillot J, Demanche C, Hugot JP, Berthelemy M, Wakefield AE, Dei-Cas E, Chermette R. Parallel phylogenies of Pneumocystis species and their mammalian hosts. J Eukaryot Microbiol. 2001;48:113–115. doi: 10.1111/j.1550-7408.2001.tb00475.x. [DOI] [PubMed] [Google Scholar]
- Demanche C, Berthelemy M, Petit T, Polack B, Wakefield AE, Dei-Cas E, Guillot J. Phylogeny of Pneumocystis carinii from 18 primate species confirms host specificity and suggests coevolution. J Clin Microbiol. 2001;39:2126–2133. doi: 10.1128/JCM.39.6.2126-2133.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Demanche C, Barriel V, Dei-Cas E, Guillot J. Phylogenetic systematics and evolution of primate-derived Pneumocystis based on mitochondrial or nuclear DNA sequence comparison. Syst Biol. 2003;52:735–744. doi: 10.1080/10635150390250893. [DOI] [PubMed] [Google Scholar]
- Akbar H, Pinçon C, Aliouat CM, Derouiche S, Taylor ML, Pottier M, Carreto-Binaghi LH, González-González A, Courpon A, Barriel V, Guillot J, Chabé M, Suarez-Alvarez RO, Aliouat EM, Dei-Cas E, Demanche C. Characterizing Pneumocystis in the lungs of bats: understanding Pneumocystis evolution and the spread of Pneumocystis organisms in mammal populations. Appl Environ Microbiol. 2012;78:8122–8136. doi: 10.1128/AEM.01791-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabé M, Herbreteau V, Hugot JP, Bouzard N, Deruyter L, Morand S, Dei-Cas E. Pneumocystis carinii and Pneumocystis wakefieldiae in wild Rattus norvegicus trapped in Thailand. J Eukaryot Microbiol. 2010;57:213–217. doi: 10.1111/j.1550-7408.2009.00465.x. [DOI] [PubMed] [Google Scholar]
- Derouiche S, Deville M, Taylor ML, Akbar H, Guillot J, Carreto-Binaghi LE, Pottier M, Aliouat EM, Aliouat-Denis CM, Dei-Cas E, Demanche C. Pneumocystis diversity as a phylogeographic tool. Mem Inst Oswaldo Cruz. 2009;104:112–117. doi: 10.1590/s0074-02762009000100017. [DOI] [PubMed] [Google Scholar]
- Gannon WL, Sikes RS. the Animal Care and Use Committee of the American Society of Mammalogists. Guidelines of the American Society of Mammalogists for the use of wild mammals in research. J Mammal. 2007;88:809–823. doi: 10.1093/jmammal/gyw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek R, Feucht A, Aepinus C, Just-Nubling G, Robertson VJ, Knobloch J, Hohle R. Evaluation of two nested PCR assays for detection of Histoplasma capsulatum DNA in human tissue. J Clin Microbiol. 2002;40:1644–1647. doi: 10.1128/JCM.40.5.1644-1647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield AE, Pixley FJ, Banerji S, Sinclair K, Millar RF, Moxon ER, Hopkin JM. Amplification of mitochondrial ribosomal RNA sequences from Pneumocystis carinii DNA of rat and human origin. Mol Biochem Parasitol. 1990;43:69–76. doi: 10.1016/0166-6851(90)90131-5. [DOI] [PubMed] [Google Scholar]
- Wakefield AE. DNA sequences identical to Pneumocystis carinii f. sp. carinii and Pneumocystis carinii f. sp. hominis in samples of air spora. J Clin Microbiol. 1996;34:1754–1759. doi: 10.1128/jcm.34.7.1754-1759.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolaki AG, Beckers P, Wakefield AE. Pre-AIDS era isolates of Pneumocystis carinii f. sp. hominis: high genotypic similarity with contemporary isolates. J Clin Microbiol. 1998;36:90–93. doi: 10.1128/jcm.36.1.90-93.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor ML, Ruíz-Palacios GM, Reyes-Montes MR, Rodríguez-Arellanes G, Carreto-Binaghi LE, Duarte-Escalante E, Hernández-Ramírez A, Pérez A, Suárez-Álvarez RO, Roldán-Aragón Y, Romero-Martínez R, Sahaza-Carmona JH, Sifuentes-Osornio J, Soto-Ramírez LE, Peña-Sandoval G. Identification of the infection source of an unusual outbreak of histoplasmosis, in a hotel in Acapulco, state of Guerrero, Mexico. FEMS Immunol Med Microbiol. 2005;45:435–441. doi: 10.1016/j.femsim.2005.05.017. [DOI] [PubMed] [Google Scholar]
- Ceballos G, Oliva G. Los Mamíferos Silvestres de México. FCE-CONABIO: México DF; 2005. [Google Scholar]
- Cox FEG. Concomitant infections, parasites and immune responses. Parasitology. 2001;122:23–38. doi: 10.1017/s003118200001698x. [DOI] [PubMed] [Google Scholar]
- Tschudy J, Michail S. Disseminated histoplasmosis and pneumocystis pneumonia in a child with Crohn disease receiving infliximab. J Pediatr Gastroenterol Nutr. 2010;51:221–222. doi: 10.1097/MPG.0b013e3181c2c10d. [DOI] [PubMed] [Google Scholar]
- De Lima IS, Lima AKF, Morishi MO, Salem JI, Braga De Souza JV. Selection and optimization of PCR-based methods for the detection of Histoplasma capsulatum var. capsulatum. Rev Iberoam Micol. 2012;29:34–39. doi: 10.1016/j.riam.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Espinosa-Avilés D, Taylor ML, Reyes-Montes MR, Pérez-Torres A. Molecular findings of disseminated histoplasmosis in two captive snow leopards (Uncia uncia) J Zoo Wildl Med. 2008;39:450–454. doi: 10.1638/2006-0062.1. [DOI] [PubMed] [Google Scholar]
- Frías De León MG, Arenas-López G, Taylor ML, Acosta-Altamirano G, Reyes-Montes MR. Development of specific sequence characterized amplified region markers for detecting Histoplasma capsulatum in clinical and environmental samples. J Clin Microbiol. 2012;50:3673–3679. doi: 10.1128/JCM.05271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Mirdha BR, Guleria R, Mohan A, Agarwal SK, Kumar L, Kabra SK, Samantaray JC. Improved detection of Pneumocystis jirovecii infection in a tertiary care reference hospital in India. Scand J Infect Dis. 2007;39:571–576. doi: 10.1080/00365540601131976. [DOI] [PubMed] [Google Scholar]
- Morgan GS, Ridgway RB. Late Pliocene (late Blancan) vertebrates from the St. Petersburg times site, Pinellas County, Florida, with a brief review of Florida Blancan faunas. Florida Paleontol. 1987;1:1–22. [Google Scholar]
- Scott P, Keely SP, Fischer JM, Stringer JR. Evolution and speciation of Pneumocystis. J Eukaryot Microbiol. 2003;50:624–626. doi: 10.1111/j.1550-7408.2003.tb00655.x. [DOI] [PubMed] [Google Scholar]


