Abstract
Triosephosphate isomerase 1 (TPI1) is a member of the glycolytic pathway, which is a critical source of energy for motility in mouse sperm. By immunoblotting, we detected two male, germ line-specific TPI1 bands (Mr 33,400 and 30,800) as well as the somatic-type band (Mr 27,700). Although all three bands were observed in spermatogenic cells, somatic-type TPI1 disappeared from sperm during epididymal maturation. In vitro dephosphorylation analysis suggested that the two male, germ line-specific TPI1 bands were not the result of phosphorylation of the 27,700 Mr TPI1 band. The Mr 33,400; 30,800; and 27,700 TPI1 bands corresponded to the respective sizes of the proteins predicted to use the first, second, and third possible initiation codons of the Tpi1 cDNA. We performed immunofluorescence on epididymal sperm and determined that TPI1 specifically localized in the principal piece. The antibody staining was stronger in cauda epididymal sperm than in caput epididymal sperm, a finding consistent with the identification of TPI1 as a cauda epididymal sperm-enriched protein. Immunofluorescence with sodium dodecyl sulfate (SDS)-insoluble flagellar accessory structures showed a strong TPI1 signal only in the principal piece, indicating that TPI1 is a component of the fibrous sheath. Northern blot hybridization detected longer Tpi1 transcripts (1.56 kb) in mouse testis, whereas somatic tissues had shorter transcripts (1.32 kb). As there is only one triosephosphate isomerase gene in the mouse genome, we conclude that the three variants we see in sperm result from the use of alternative translation start codons in spermatogenic cells. Mol. Reprod. Dev. 80: 862–870, 2013. © 2013 The Authors. Published by Wiley Periodicals, Inc. This is an open access article under the terms of the Creative Commons Attribution-NonCommercial-NoDerivs License, which permits use and distribution in any medium, provided the original work is properly cited, the use is non-commercial and no modifications or adaptations are made.
Keywords: sperm, fibrous sheath, flagellum, triosephosphate isomerase, spermatogenesis
INTRODUCTION
Male germ cells have to produce ATP from available substrates for their cellular activity and for differentiation into spermatozoa. Although spermatocytes and spermatids prefer oxidative phosphorylation as a means for ATP production (Nakamura et al., ,), studies have suggested that sperm of many mammals switch to glycolysis for ATP production during epididymal maturation (Storey and Kayne, 1975). During spermatogenesis, a number of somatic-type glycolytic enzymes are replaced by male germ cell-specific isozymes, many of which are found in association with the fibrous sheath of the flagellar principal piece (McCarrey and Thomas, 1987; Sakai et al., 1987; Welch et al., 1992a; Mori et al., 1993).
Glycolysis clearly plays an essential role as an energy pathway to fuel basal motility in mouse sperm, as is evident by studies of male mice with genetic deletions of the sperm-specific forms of key glycolytic enzymes, such as glyceraldehyde 3-phosphate (GAP) dehydrogenase-S, lactate dehydrogenase C, and phosphoglycerate kinase-2 (Miki et al., 2004; Odet et al., 2008; Danshina et al., 2010). When oxidative phosphorylation is suppressed by carbonyl cyanide m-chlorophenylhydrazone, sperm motility and ATP production are not affected; on the other hand, sperm motility and ATP content are negatively impacted when glucose is replaced in the medium by a non-hydrolyzable analog, 2-deoxyglucose, even though pyruvate or lactate is also present to fuel mitochondrial respiration (Mukai and Okuno, 2004).
Glycolysis converts glucose into pyruvate via a series of reactions by ten enzymes, yielding two ATPs from a single glucose molecule. During glycolysis, aldolase converts fructose 1,6-diphosphate to GAP and dihydroxyacetone phosphate (DHAP). Only GAP is a direct substrate for GAP dehydrogenase, thus triosephosphate isomerase 1 (TPI1) is required to convert DHAP to GAP. The conversion reaction is highly efficient, and is also very reversible, forming DHAP from GAP in cells involved in gluconeogenesis. TPI1 has been found in most organisms, and has been studied in the human and rat male germ lines (Russell and Kim, 1996; Auer et al., 2004); yet, little is known about the mouse enzyme. We previously reported that mouse TPI1 was present in the sperm flagellar accessory structures and was also identified as a cauda epididymal sperm-enriched protein (Cao et al., 2006; Ijiri et al., 2011). The glycolytic enzyme aldolase has male germ line-specific isozymes (ALDOA, ALDOAV2, ALDOART1, ALDOART2), and anti-muscle aldolase or anti-ALDOA antibodies detected the ubiquitous shorter (Mr 39,000) and longer (Mr 50,000) male germ cell-specific isozymes (Vemuganti et al., 2007; Arcelay et al., 2008; Ijiri et al., 2011). Similar to aldolase, immunoblotting showed two male germ line-specific Mr 33,400 and 30,800 TPI1 bands as well as the somatic-type Mr 27,700 band (Ijiri et al., 2011).
In this study, we investigated mouse TPI1 further by analyzing protein distribution in somatic and reproductive tissues. We also examined the localization of this protein during epididymal maturation using caput and cauda epididymal sperm. As it is reported that multiple glycolytic enzymes are tightly bound to the fibrous sheath in mouse (Krisfalusi et al., 2006), we also performed immunofluorescence with sodium dodecyl sulfate (SDS)-insoluble flagellar accessory structures to determine if TPI1 was specifically localized in the fibrous sheath. We demonstrated that in the mouse, TPI1 existed as three isozymes (two male germ line-specific and one somatic tissue type). The male germ line-specific isozymes were present in spermatogenic cells and epididymal sperm, and associate with the caput and cauda epididymal sperm flagellum. Furthermore, the mouse Tpi1 gene sequence and transcripts were examined by Northern blot hybridization analyses and Rapid Amplification of cDNA Ends (RACE), respectively, to elucidate how TPI1 isozymes were produced in male mouse germ cells. We present evidence that the three variants present in sperm result from the use in spermatogenic cells of alternative translation start codons for Tpi1, the only triosephosphate isomerase gene in the mouse genome.
RESULTS
The Male Mouse Germ Line Exhibits Two Specific TPI1 Isozymes
To determine where TPI1 was expressed and whether or not a specific isozyme(s) existed, we performed SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and immunoblotting with various somatic and reproductive tissues. The somatic-type TPI1 protein (Mr 27,700) was detected in somatic tissues, ovary, and testis (Fig. 1). In spermatogenic cells and sperm, we found two male germ line-specific TPI1 bands (Mr 33,400 and 30,800). Although all three bands were observed in germ cells from pachytene spermatocytes to caput epididymal sperm, somatic-type TPI1 disappeared from sperm during epididymal maturation. The two male germ line-specific bands increased gradually in relative amounts, becoming most abundant in cauda epididymal sperm.
Figure 1.
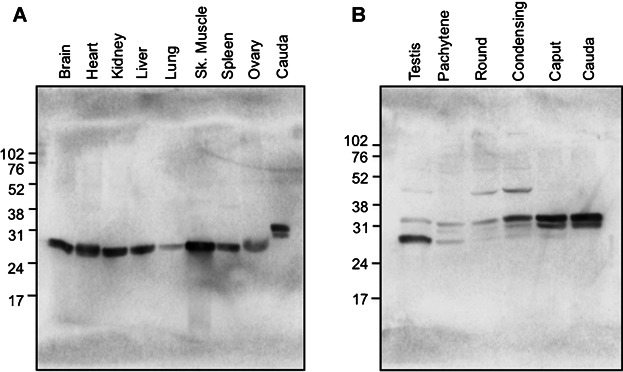
SDS–PAGE and immunoblotting of protein extracts from somatic tissues, reproductive tissues, and spermatogenic cells. Immunoblotting with protein extracts from brain, heart, kidney, liver, lung, skeletal muscle, spleen, ovary, and cauda epididymal sperm (A). Immunoblotting with protein extracts from testis, pachytene spermatocytes, round spermatids, condensing spermatids, caput epididymal sperm, and cauda epididymal sperm (B). The somatic-type TPI1 protein (Mr 27,700) was detected in all protein samples examined, except cauda epididymal sperm, using an anti-TPI1 antibody. In spermatogenic cells and sperm, two male germ line-specific TPI1 bands (Mr 33,400 and 30,800) were present.
The Male Germ Line-Specific TPI1 Isozymes Do Not Result From Differential Phosphorylation
Calf intestinal alkaline phosphatase (CIP) was used to determine if the differences in relative mobility of the three TPI1 isozymes were due to differential phosphorylation. Proteins extracted from testis and mature epididymal sperm were treated with alkaline phosphatase and then examined by immunoblotting with anti-TPI1 antibody. As a positive control, cauda epididymal sperm proteins were probed with anti-AKAP4 antibody with and without phosphatease treatment to demonstrate a shift in mobility caused by dephosphorylation (Johnson et al., 1997). In vitro dephosphorylation analysis did not produce any change in the TPI1 relative mobility, indicating that the two male germ line-specific TPI1 isozymes did not result from differences in phosphorylation status (Fig. 2).
Figure 2.
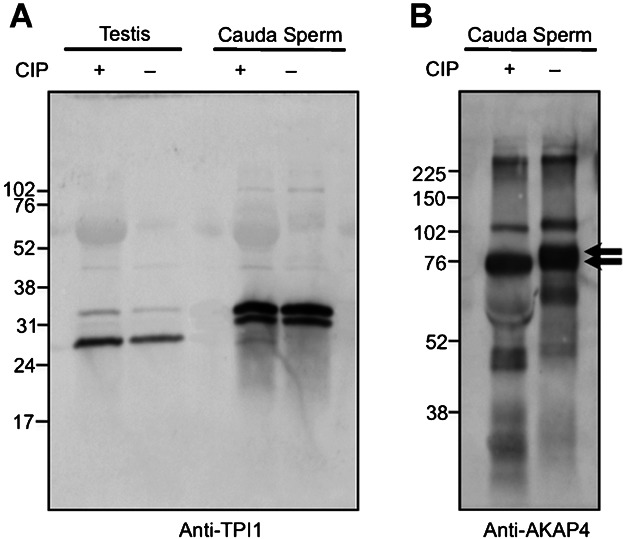
In vitro dephosphorylation of TPI1 in mouse sperm. CIP treatment of testis and cauda epididymal sperm proteins probed with anti-TPI1 antibody (A) and cauda epididymal sperm proteins probed with anti-AKAP4 antibody as a positive control (B). The two arrows point to the two AKAP4 forms present prior to dephosphorylation. In vitro dephosphorylation analysis indicated that the two male germ line-specific TPI1 isozymes did not result from differences in phosphorylation.
Northern Blot Hybridization Detects Longer Tpi1 Transcripts in the Mouse Testis
To determine if the existence of male germ line-specific Tpi1 transcripts could explain the differences seen in the proteins extracted from spermatogenic cells, we performed Northern blot hybridization on messenger RNA from brain, heart, kidney, ovary, and testis. A single band at 1.32 kb was noted in brain, heart, kidney, and ovary (Fig. 3). In the testis, a larger but broader single band was also observed. The center of this band corresponded to a size of 1.56 kb.
Figure 3.
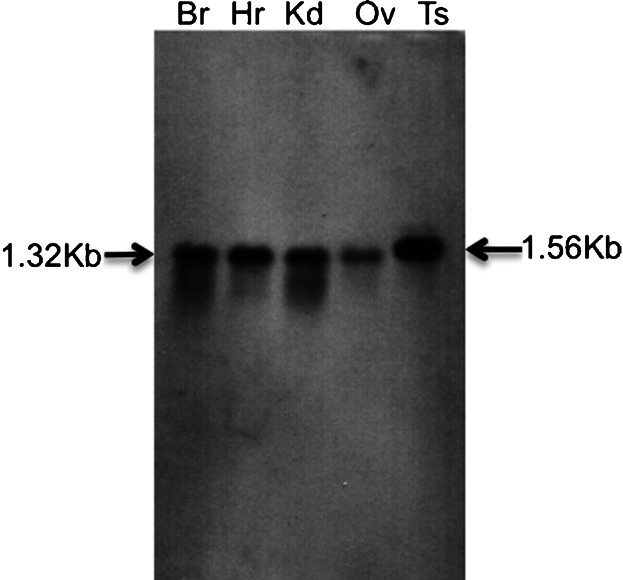
Northern blot hybridization with a Tpi1 antisense probe. Northern blot hybridization was performed to confirm how many male germ line-specific Tpi1 bands were present. Message RNA from brain (Br), heart (Hr), kidney (Kd), ovary (Ov), and testis (Ts) were examined. This method detected a longer, testis-specific Tpi1 transcript of 1.56 kb; somatic tissues and ovary contained only a 1.32-kb transcript.
The Male Mouse Germ Line Expresses a Spermatogenic-Specific Tpi1 mRNA Variant
Recognizing that the larger-sized mRNAs in the testis could result from extension at either end of the message, cDNAs were created and 5′ RACE was performed to identify potential initiation codons. Two database cDNA sequences exist for mouse Tpi1. The original sequence, NM_009415.1, was based upon predictions from the genomic sequence on mouse chromosome 6 and has subsequently been replaced by an updated, predicted sequence, NM_009415.2, which lacks the 5′-most in-frame, upstream stop codon that is present in NM_009415.1. Using 5′ RACE, we found two different transcripts in mouse spermatogenic cell cDNA. Transcript 1 contained all three potential initiation codons and an in-frame, upstream stop codon, whereas, transcript 2, like NM_009415.2, lacked the in-frame, upstream stop codon and the first potential initiation codon (Suppl. Fig. S1).
In rat, testicular Tpi1 mRNA contains an additional 100 bases in the 3′-untranslated region (UTR) that is specific to male germ cells (Russell and Kim, 1996). We tested if mouse Tpi1 mRNA contained a region similar to the 3′ UTR of rat Tpi1 using 3′ RACE. The predicted mouse Tpi1 sequence, NM_009415.2, includes the 3′-UTR extension that is homologous to the germ line-specific 3′-UTR of rat Tpi1 cDNA; however, our 3′-RACE analyses did not detect the extended sequence (Suppl. Fig. S2). Sequences from ten different 3′-RACE clones terminated at the same point predicted by NM_009415.1, indicating that unlike the rat, the mouse does not express a male germ line-specific 3′-UTR. The nucleotide sequences did contain a canonical polyadenylation signal for Tpi1 (Suppl Fig. S2, box).
TPI1 Isozymes Likely Result From Three Different Initiation Codons
To determine the basis for the differences in Mr of the germ-cell forms of TPI1, open reading frames from the full-length mouse Tpi1 cDNA were predicted based on our 5′-RACE results and NM_009415.1. Three potential initiation codons were identified in the Tpi1 cDNA. The Mr 33,400; 30,800; and 27,700 TPI1 bands corresponded to the sizes of the proteins predicted to use the first (299 amino acids long, calculated molecular weight: 32,192), second (286 amino acids, calculated molecular weight: 30,890), and third (249 amino acids, calculated molecular weight: 26,712) initiation codons of the Tpi1 cDNA, respectively (Suppl. Fig. S3; the two orange boxes indicate the putative translation initiation codons of the two male germ line-specific TPI1 isozymes, and the green box is the known translation initiation codon of somatic-type TPI1). An in-frame stop codon is located 42 bp upstream from the first putative start codon at nucleotide 141 (red box). The second and third initiation codons contain a strong Kozak consensus sequence following the (gcc)gccRccAUGG sequence, consisting of a purine in position −3 and a guanine in position +4 (Kozak, 1987). The first initiation codon has a weak Kozak consensus sequence, and does not contain a purine in position −3.
Germ Line-Specific TPI1 Isozymes Partition to the Flagellar Accessory Structures
To assess if TPI1 associated with components of the flagellar accessory structures such as the fibrous sheath, cauda epididymal sperm proteins were fractionated by extracting with S-EDTA (1% SDS, 75 mM NaCl, 10 mM EDTA, pH 6.0), 0.1% Triton X-100, or 0.1% Triton X-100 followed by S-EDTA; resulting fractions were analyzed by immunoblotting with anti-TPI1 antibody. S-EDTA solubilizes these components and dissolves the axoneme, leaving behind flagellar accessory structures such as the fibrous sheath, outer dense fibers, and remnants of the mitochondrial sheath. Triton X-100, on the other hand, solubilizes membranes and releases soluble, cytoplasmic components but leaves the axoneme intact. The two male germ line-specific bands of Mr 33,400 and 30,800 were enriched in the S-EDTA-treated, sperm-tail fraction (Fig. 4). The upper TPI1 band (Mr 33,400) was faintly observed in the sperm head fraction. The two germ line-specific isozyme bands were detected in Triton X-100/S-EDTA insoluble sperm proteins, but not in the soluble fraction.
Figure 4.
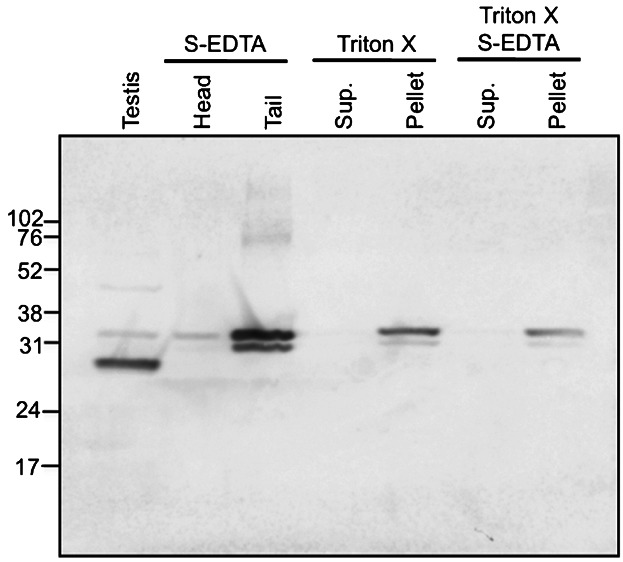
SDS–PAGE and immunoblotting of detergent-extracted sperm proteins. Immunoblotting on SDS-insoluble head and tail proteins, Triton X-100-soluble and -insoluble, whole sperm proteins, and Triton X-100/S-EDTA-soluble and -insoluble whole sperm proteins was performed. The two male germ line-specific bands (Mr 33,400 and 30,800) were enriched in the S-EDTA-treated sperm tail fraction, in Triton X-100-insoluble sperm proteins, and in Triton X-100/S-EDTA-insoluble sperm proteins.
TPI1 Localizes to the Principal Piece of Mouse Epididymal Sperm
Indirect immunofluorescence was performed on caput and cauda epididymal sperm treated with anti-TPI1 antibody (Fig. 5), then exposed to fluorescence (left panels) and Nomarski differential interference contrast (right panels) microscopy. TPI1 specifically localized to the principal piece of caput epididymal sperm (Fig. 5A) and cauda epididymal sperm (Fig. 5B). After solubilization of sperm with S-EDTA, TPI1 remained detectable in the principal piece of the SDS-insoluble flagellar accessory structures (Fig. 5C). Fluorescence appeared to be associated with the longitudinal columns of the fibrous sheath. No signal was observed in normal-IgG controls (data not shown).
Figure 5.
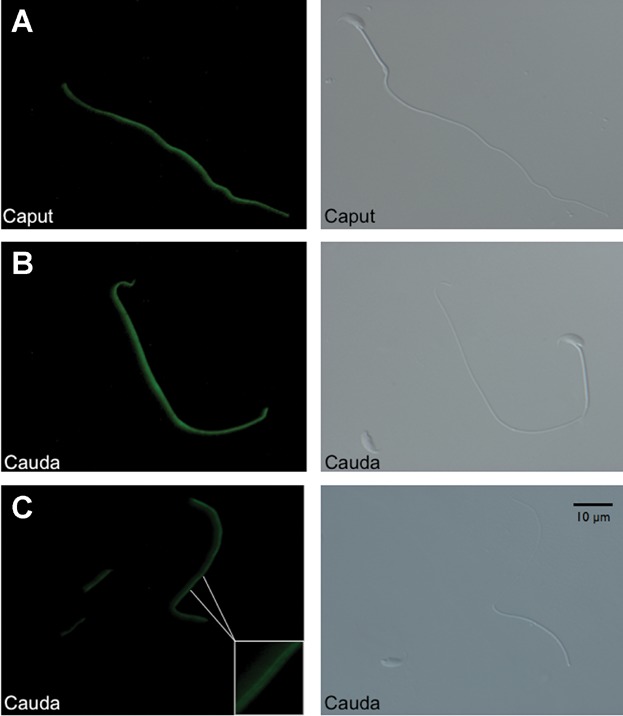
Indirect immunofluorescence with anti-TPI1 antibody. TPI1 was detected in the principal piece of caput epididymal sperm (A), cauda epididymal sperm (B), and SDS-insoluble tail accessory structures of cauda epididymal sperm (C). Scale bar, 10 µm.
DISCUSSION
Glycolysis plays an important role in energy production with mammalian sperm. All glycolytic enzymes play key roles, and the requirement for TPI1 in the male germ cell is evident from inhibitor studies, which demonstrated that a TPI1 inhibitor decreases rat sperm motility by 50%, consistent with the hypothesis that the conversion of DHAP to GAP increases the efficiency of the glycolytic process (Bone and Cooper, 2000; Bone et al., 2001). Although some information is available concerning TPI1 in sperm from a limited number of species, an extensive analysis of the isoforms present and the localization of the protein in sperm have not been published. We previously reported that mouse TPI1 was present in the sperm flagellar accessory structures and was identified as a cauda epididymal sperm-differential protein by proteomic analysis (Cao et al., 2006; Ijiri et al., 2011). Here, we have shown that TPI1 is restricted to the principal piece of the sperm flagellum, and is tethered to the fibrous sheath.
Somatic TPI1 was present in pachytene spermatocytes, but declined in amount as spermatogenic cells matured. Conversely, late-meiotic and post-meiotic male germ cells expressed two specific TPI1 isozymes (Mr 33,400 and 30,800; Fig. 1). During epididymal maturation, the two male germ line-specific TPI1 isozymes were enriched while the somatic-type disappeared. Cauda epididymal sperm exhibited only the two male germ line-specific TPI1s, suggesting that these isozymes are important for sperm function. Loss of the somatic form during spermatogenesis and epididymal maturation probably reflects remodeling of the male germ cell through the shedding of excess cytoplasm in residual bodies and cytoplasmic droplets from spermatids and cauda epididymal sperm, respectively.
From our studies, we conclude that the Mr 33,400; 30,800; and 27,700 TPI1 isozymes corresponded to the sizes of the proteins predicted to use the respective first, second, and third possible initiation codons of the Tpi1 cDNA (Suppl. Fig. S3). The methionine present at the third possible initiation codon is reportedly used to generate somatic TPI1 (Yuan et al., 1979; Cheng et al., 1990). The use of alternative start codons is similar to the gonadotropin-regulated testicular RNA helicase gene, which results from the utilization of optional translation initiation codons in the rat testis (Sheng et al., 2003). Northern blot hybridization for Tpi1 mRNA detects longer Tpi1 transcripts in mouse testis compared to somatic tissue or female reproductive tissues (Fig. 3), and is independent. Additionally, the differences in relative mobilities of the germ line-specific TPI1 proteins were not due to differential phosphorylation (Fig. 2). The expression of sperm-specific glycolytic enzymes from the same gene that is used in somatic cells contrasts to the situation with other glycolytic enzymes. In the cases of lactate dehydrogenase and phosphoglycerate kinase, alternate genes are responsible for the sperm-specific isozymes (Vemuganti et al., 2010). These genes usually originate from retrotransposition events. In the case of triosephosphate isomerase, retrotransposed sequences are present in the human, but these sequences do not appear to be expressed in the testis (Vemuganti et al., 2010). At present, the mechanism regulating the use of alternative start codons in male germ-cell TPI1 isozyme translation is unknown.
Amino terminal extensions are common for proteins that associate with the fibrous sheath in a detergent-resistant manner. Examples of this phenomenon include germ cell-specific hexokinase 1 and GAP dehydrogenase (Welch et al., 1992b; Mori et al., 1998; Travis et al., ,). The male germ line-specific N-termini of TPI1, which contain additional 50 and 37 amino acids, are not predicted to have an alpha helical structure (data not shown) as do the tethering domains of the regulatory subunits of protein kinase A (Visconti et al., 1997; Miki and Eddy, 1999). To examine this issue further, we determined whether or not these isozymes associated with the fibrous sheath. S-EDTA solubilizes and dissolves the axoneme, leaving behind flagellar accessory structures such as the fibrous sheath, outer dense fibers, and remnants of the mitochondrial sheath. Triton X-100 solubilizes membranes and releases soluble cytoplasmic components, but leaves the axoneme intact. The germ line-specific TPI1 bands (Mr 33,400 and 30,800) were detected in the S-EDTA-treated sperm tail fraction and Triton X-100/S-EDTA insoluble sperm proteins, but not in S-EDTA-treated head and Triton X-100/S-EDTA-soluble sperm protein fractions (Fig. 4). Using indirect immunofluorescence with anti-TPI1 antibody, we showed that the enzyme localized to the principal piece of mouse epididymal sperm, and appeared to be present in the longitudinal columns of the fibrous sheath in cauda epididymal sperm (Fig. 5). These results are consistent with studies of other glycolytic enzymes that were found to be tethered to the fibrous sheath of the sperm flagellum (Krisfalusi et al., 2006; Nakamura et al., 2013).
The studies presented here provide evidence for the attachment of another glycolytic enzyme to the fibrous sheath. Together with hexokinase, aldolase, phosphoglycerate kinase, GAP dehydrogenase, enolase, and pyruvate kinase, TPI1 associates with the fibrous sheath to form a specialized “metabolosome” within the sperm flagellum. This compartmentalized and particulate structure forms the primary mechanism for generating ATP within a cell with very little cytoplasm. Additional studies should be performed to define the complete metabolic pathways within the sperm, including the further processing of pyruvate and lactate. Such studies may discover new approaches for contraceptive development. In addition, the sperm flagellum serves as a model for the development of micro- and nanoscale hybrid material systems that can attach biological components, such as enzymes, to inorganic surfaces without compromising catalytic function (Mukai et al., 2013).
MATERIALS AND METHODS
Spermatogenic Cell Isolation
All animal procedures were approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Mixed germ cells were prepared from decapsulated testes of adult male mice (Crl:CD1(ICR) retired breeders; Charles River Laboratories, Wilmington, MA) by sequential dissociation with collagenase and trypsin-DNase I (Bellvé et al., 1977). To purify populations of pachytene spermatocytes, round spermatids, and condensing spermatids, mixed germ cells were separated at unit gravity in a 2–4% bovine serum albumin gradient in Eagle's Essential Medium with Earle's Salts (Romrell et al., 1976; Gerton and Millette, 1984). Both the pachytene spermatocyte and round spermatid populations were at least 85% pure, as determined by microscopic examination and differential counting with a hemocytometer; the condensing spermatid population was approximately 40–50% pure, with the balance primarily composed of anucleate residual bodies and round spermatids.
Purification of Caput and Cauda Epididymal Sperm
Sperm were collected from the caput and caudae epididymides of male mice by cutting the epididymides and extruding the sperm at 37°C into phosphate-buffered saline (PBS; 2.68 mM KCl, 136.09 mM NaCl, 1.47 mM KH2PO4, 8.07 mM Na2HPO4, pH 7.4) containing protease inhibitors (Roche Applied Science, Indianapolis, IN). Caput epididymal sperm were purified by centrifugation at 400g for 20 min at room temperature through a 35% SupraSperm 100 solution (MidAtlantic Devices, Mt. Laurel, NJ) in PBS. Purified sperm were collected from the pellet, resuspended in PBS at 4°C, counted, and assessed for purity. Cauda epididymal sperm were collected by centrifugation at 800g for 5 min at room temperature, resuspended in PBS at 4°C, and counted.
Purification of SDS-Insoluble Sperm Heads and Tails
Cauda epididymal sperm were homogenized in S-EDTA, layered onto a 1.6 M sucrose cushion in S-EDTA, and centrifuged at 5,000g for 1 hr at room temperature (O'Brien and Bellvé, 1980). The SDS-resistant head structures (nuclei) were collected from the pellet. The SDS-resistant tail structures (flagellar accessory structures lacking the plasma membrane and axonemes) were collected from the interface. The purity was visually assessed by light microscopy to determine adequate head and flagellar accessory structure (“tail”) separation.
Detergent Fractionation of Sperm Cells
Cauda epididymal sperm were collected as described above. One million cells in 100 µl of PBS containing 0.1% Triton X-100 were incubated at room temperature for 10 min, then centrifuged at 10,000g for 1 min. Triton X-100-soluble and -insoluble proteins were collected from the supernatant and pellet, respectively. Further detergent extraction was performed on the Triton X-100-insoluble proteins (pellets). One hundred microliters of S-EDTA was added to the Triton X-100-insoluble protein pellet, homogenized, incubated at room temperature for 10 min, and centrifuged at 10,000g for 1 min. The Triton X-100/S-EDTA-soluble and -insoluble proteins were collected from the supernatant and pellet, respectively.
In Vitro Dephosphorylation by Alkaline Phosphatase
CIP (New England Biolabs, Ipswich, MA) can be used to release phosphate groups from phosphorylated tyrosine, serine, and threonine residues (Johnson et al., 1997). Proteins were dephosphorylated according to manufacturer's instructions. Briefly, 1.75–3.5 U of CIP/µg protein was incubated with protease inhibitors at 37°C for 30 min. CIP-treated proteins were then subjected to immunoblotting with an anti-TPI1 (10713-1-AP, rabbit polyclonal antibody; ProteinTech Group, Chicago, IL) or anti-AKAP4 antibody, as described below. The antibody against AKAP4, originally assigned as anti-AKAP82, has been previously characterized (Carrera et al., 1994).
Protein Extraction and Immunoblot Analysis
The spermatogenic cells and sperm cells were concentrated by centrifugation, washed in 1 ml of PBS, resuspended in sample buffer (58 mM Tris–HCl, pH 6.8, 1.7% SDS, 6% glycerol, 100 mM dithiothreitol [DTT]), and boiled for 5 min. Detergent-extracted proteins were mixed with sample buffer and boiled for 5 min. After centrifugation, the supernatants were recovered and saved; protein concentrations were determined by the BCA Protein Assay Kit-Reducing Agent Compatible (Pierce Chemical Co., Rockford, IL). Other tissues were homogenized, sonicated, and boiled in sample buffer without DTT. After centrifugation, the supernatants were recovered and saved. Protein concentrations were determined by the Micro BCA Protein Assay Kit (Pierce Chemical Co.) and then DTT and bromophenol blue were added to final concentrations of 100 mM and 0.002%, respectively. The samples were boiled for 5 min, and protein samples (10 or 20 µg/lane) were separated by SDS–PAGE in 10% or 12% polyacrylamide gels for AKAP4 or TPI1, respectively (Laemmli, 1970). The gels were then transferred to polyvinylidene difluoride membranes (Towbin et al., 1979). After the membranes were blocked with TBST (125 mM NaCl, 25 mM Tris–HCl, pH 8.0, 0.1% Tween-20) containing 5% nonfat dry milk, they were incubated with primary antibody for 1 hr (1.05 µg/ml anti-TPI1 or 1:10,000 anti-AKAP4). After washing with TBST, the blots were incubated for 1 hr with secondary antibody (1:5,000 alkaline phosphate-conjugated goat anti-rabbit IgG whole antibody [GE Healthcare, Milwaukee, WI] in 5% nonfat dry milk in TBST). After washing again with TBST, the bound enzyme was developed with Enhanced Chemifluorescence Western Blotting Reagent Pack (GE Healthcare) according to the manufacturer's directions. Images were produced using a Storm 860 scanner (GE Healthcare). Negative controls using normal sera or secondary antibody-alone were also used to check for specificity.
Indirect Immunofluorescence Analysis
One million caput or cauda epididymal sperm, collected as described above, were attached to polylysine-coated coverslips for 15 min, fixed with 4% paraformaldehyde in PBS for 15 min, and then permeabilized with methanol for 2 min at −20°C. Permeabilization with methanol was omitted when preparing S-EDTA-treated cauda epididymal sperm. After extensive washing with PBS, the coverslips were incubated at 4°C overnight with 10% goat serum in PBS (blocking solution). The following day, coverslips were incubated at 37°C for 1 hr with primary antibody (2.1 µg/ml anti-TPI1) diluted in blocking solution. After washing with PBS, the coverslips were incubated for 1 hr at 37°C with Alexa Fluor 488-conjugated goat anti-rabbit IgG (H + L) secondary antibodies (1:500; Life Technologies, Carlsbad, CA) diluted in blocking solution. Finally, the coverslips were mounted on slides using 15 µl of Fluoromount-G (Southern Biotechnology Associates, Birmingham, AL), observed with a Nikon Eclipse TE 2000-U inverted microscope (Nikon Instruments, Melville, NY), and photographed with a CFW-1310C digital FireWire camera (Scion, Frederick, MD) using the NIH ImageJ imaging software available online (http://rsb.info.nih.gov/ij/). Nomarski differential interference contrast micrographs were photographed in parallel with the fluorescence images. Negative controls using normal sera or secondary antibody-alone were used to check for specificity.
Preparation of RNA
Messenger RNAs were prepared from brain, heart, kidney, testis, and ovary using Dynabeads mRNA Direct Kit (Invitrogen, Grand Island, NY) according to manufacturer's instructions for Northern blot hybridization. Total RNA from mixed germ cells was used for 5′- and 3′-RACE analyses.
Northern Blot Hybridization
Northern blot hybridization was performed similarly to a previous method (Cao et al., 2011). Messenger RNAs (0.5 µg each) from brain, heart, kidney, testis, and ovary were separated by electrophoresis in a 1% agarose gel, containing formaldehyde, and transferred to Hybond N+ membranes (GE Healthcare). The probes were derived from an initial PCR product (Tpi1 [accession number: NM_009415] F: 5′-TGACCTTCAGAGACTTGAGCC and R: 5′-CGGTGGGAGCAGTTACTAAAC; product size: 851 bp) and ligated into a pCRII-TOPO vector (Invitrogen). The probe was sequenced, then quantified by comparison to a Hind III ladder (New England Biolabs) and labeled with digoxigenin (DIG)-dUTP using in vitro transcription (DIG Northern Starter Kit, Roche). The probe was used for hybridization to Hybond N+ membranes at 68°C for 2 hr. Membranes were washed to a stringency of 1× SSC (diluted from 20× SSC: 3 M sodium chloride, 0.3 M sodium citrate, pH 7.0), then 0.05× SSC and 0.1% SDS up to 68°C. Immunological detection with anti-DIG antibody and chemiluminescence reaction with CDP-Star were used to assess the presence of Tpi1 RNA following the manufacturer's instructions.
5′- and 3′-Rapid Amplification of cDNA Ends, Cloning of Tpi1, and cDNA Sequence Analyses
Mixed germ cell amplicons, produced as described above, were cloned into a pCR2.1-TOPO vector (Invitrogen). Plasmid DNA was prepared and sequenced as directed by the manufacturer's instructions. The initial primer designs were based on the Mus musculus Tpi1 mRNA sequence (NM_009415). The RLM-RACE kit (Ambion, Austin, TX) was used for 5′- and 3′-RACE, and 1 µg of total RNA aliquots were used as templates for reverse transcription with the supplied 5′- (5′-GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAAA) and 3′- (5′-GCGAGCACAGAATTAATACGACTCACTATAGGT-12VN) RACE adapters. The cDNA was then subjected to PCR using the 5′- (5′-GCTCATGGCGATGAATGAACACTG) and 3′- (5′-GCGAGCACAGAATTAATACGACT) RACE primers, which were complementary to the anchored adapter, and a primer specific for Tpi1 (5′-CCCAAGTTCTACTGGATGTCTG). Gradient PCR was used to optimize annealing temperatures. All PCR products were sequenced in both directions. The cloned sequences were analyzed using MacVector Software v12.0.3 (MacVector, Inc., Cary, NC).
Glossary
Abbreviations:
- CIP
calf intestinal alkaline phosphatase
- DIG
digoxigenin
- DTT
dithiothreitol
- ECF
enhanced chemifluorescence
- PBS
phosphate-buffered saline
- RACE
rapid amplification of cDNA ends
- TPI1
triosephosphate isomerase 1
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher's web-site.
50 RACE of mouse mRNA. Utilization of 50 RACE identified the 50-UTR sequence of Tpi1 cDNAs from male mixed germ cells. Alignment of the four Tpi1 cDNA sequences: NM_009425.1 containing all three potential initiation codons, NM_009415.2 containing all three potential initiation codons, 50 RACE Transcript 1 containing all three potential initiation codons, and 50 RACE Transcript 2 containing only the second and third potential initiation codons.
30 RACE of mouse mRNA. Utilization of 30 RACE identified the 30-UTR sequence of Tpi1 cDNAs from male mixed germ cell populations. Alignment of the three Tpi1 cDNA sequences: NM_009425.1 is not predicted to contain a germ cell-specific 30-UTR, NM_009415.2 is predicted to contain a germ cell-specific 30-UTR, and the 30 RACE sequence. 30 RACE did not detect a predicted male germline-specific 30-UTR in mouse mixed germ cell cDNAs.
Amino acid sequences of TPI1 isoforms deduced from Tpi1 cDNA. Three putative translation initiation codons in full-length mouse Tpi1 cDNA (NM_009415.1) were discovered. The two orange boxes indicate putative translation initiation codons of the two male germ linespecific TPI1 isozymes, and the green box is the known translation initiation codon of somatic-type TPI1. The italicized numbers above the orange and green boxes indicate the amino acid lengths of the predicted proteins. The red box indicates the in-frame stop codon upstream from the start codons.
REFERENCES
- Arcelay E, Salicioni AM, Wertheimer E, Visconti PE. Identification of proteins undergoing tyrosine phosphorylation during mouse sperm capacitation. Int J Dev Biol. 2008;52:463–472. doi: 10.1387/ijdb.072555ea. [DOI] [PubMed] [Google Scholar]
- Auer J, Camoin L, Courtot A-M, Hotellier F, De Almeida M. Evidence that P36, a human sperm acrosomal antigen involved in the fertilization process is triosephosphate isomerase. Mol Reprod Dev. 2004;68:515–523. doi: 10.1002/mrd.20107. [DOI] [PubMed] [Google Scholar]
- Bellvé AR, Millette CF, Bhatnagar YM, O'Brien DA. Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J Histochem Cytochem. 1977;25:480–494. doi: 10.1177/25.7.893996. [DOI] [PubMed] [Google Scholar]
- Bone W, Cooper TG. In vitro inhibition of rat cauda epididymal sperm glycolytic enzymes by ornidazole, alpha-chlorohydrin and 1-chloro-3-hydroxypropanone. Int J Androl. 2000;23:284–293. doi: 10.1046/j.1365-2605.2000.00243.x. [DOI] [PubMed] [Google Scholar]
- Bone W, Jones AR, Morin C, Nieschlag E, Cooper TG. Susceptibility of glycolytic enzyme activity and motility of spermatozoa from rat, mouse, and human to inhibition by proven and putative chlorinated antifertility compounds in vitro. J Androl. 2001;22:464–470. [PubMed] [Google Scholar]
- Cao W, Gerton GL, Moss SB. Proteomic profiling of accessory structures from the mouse sperm flagellum. Mol Cell Proteomics. 2006;5:801–810. doi: 10.1074/mcp.M500322-MCP200. [DOI] [PubMed] [Google Scholar]
- Cao W, Ijiri TW, Huang AP, Gerton GL. Characterization of a novel tektin member, TEKT5, in mouse sperm. J Androl. 2011;32:55–69. doi: 10.2164/jandrol.109.009456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera A, Gerton GL, Moss SB. The major fibrous sheath polypeptide of mouse sperm: Structural and functional similarities to the A-kinase anchoring proteins. Dev Biol. 1994;165:272–284. doi: 10.1006/dbio.1994.1252. [DOI] [PubMed] [Google Scholar]
- Cheng J, Mielnicki LM, Pruitt SC, Maquat LE. Nucleotide sequence of murine triosephosphate isomerase cDNA. Nucleic Acids Res. 1990;18:4261. doi: 10.1093/nar/18.14.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danshina PV, Geyer CB, Dai Q, Goulding EH, Willis WD, Kitto GB, McCarrey JR, Eddy EM, O'Brien DA. Phosphoglycerate kinase 2 (PGK2) is essential for sperm function and male fertility in mice. Biol Reprod. 2010;82:136–1145. doi: 10.1095/biolreprod.109.079699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton GL, Millette CF. Generation of flagella by cultured mouse spermatids. J Cell Biol. 1984;98:619–628. doi: 10.1083/jcb.98.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijiri TW, Merdiushev T, Cao W, Gerton GL. Identification and validation of mouse sperm proteins correlated with epididymal maturation. Proteomics. 2011;11:4047–4062. doi: 10.1002/pmic.201100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LR, Foster JA, Haig-Ladewig L, Vanscoy H, Rubin CS, Moss SB, Gerton GL. Assembly of AKAP82, a protein kinase A anchor protein, into the fibrous sheath of mouse sperm. Dev Biol. 1997;192:340–350. doi: 10.1006/dbio.1997.8767. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisfalusi M, Miki K, Magyar PL, O'Brien DA. Multiple glycolytic enzymes are tightly bound to the fibrous sheath of mouse spermatozoa. Biol Reprod. 2006;75:270–278. doi: 10.1095/biolreprod.105.049684. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- McCarrey JR, Thomas K. Human testis-specific PGK gene lacks introns and possesses characteristics of a processed gene. Nature. 1987;326:501–505. doi: 10.1038/326501a0. [DOI] [PubMed] [Google Scholar]
- Miki K, Eddy EM. Single amino acids determine specificity of binding of protein kinase A regulatory subunits by protein kinase A anchoring proteins. J Biol Chem. 1999;274:29057–29062. doi: 10.1074/jbc.274.41.29057. [DOI] [PubMed] [Google Scholar]
- Miki K, Qu W, Goulding EH, Willis WD, Bunch DO, Strader LF, Perreault SD, Eddy EM, O'Brien DA. Glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme, is required for sperm motility and male fertility. Proc Natl Acad Sci USA. 2004;101:16501–16506. doi: 10.1073/pnas.0407708101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori C, Welch JE, Fulcher KD, O'Brien DA, Eddy EM. Unique hexokinase messenger ribonucleic acids lacking the porin-binding domain are developmentally expressed in mouse spermatogenic cells. Biol Reprod. 1993;49:191–203. doi: 10.1095/biolreprod49.2.191. [DOI] [PubMed] [Google Scholar]
- Mori C, Nakamura N, Welch JE, Gotoh H, Goulding EH, Fujioka M, Eddy EM. Mouse spermatogenic cell-specific type 1 hexokinase (mHk1-s) transcripts are expressed by alternative splicing from the mHk1 gene and the HK1-S protein is localized mainly in the sperm tail. Mol Reprod Dev. 1998;49:374–385. doi: 10.1002/(SICI)1098-2795(199804)49:4<374::AID-MRD4>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Mukai C, Okuno M. Glycolysis plays a major role for adenosine triphosphate supplementation in mouse sperm flagellar movement. Biol Reprod. 2004;71:540–547. doi: 10.1095/biolreprod.103.026054. [DOI] [PubMed] [Google Scholar]
- Mukai C, Gao L, Bergkvist M, Nelson JL, Hinchman MM, Travis AJ. Biomimicry enhances sequential reactions of tethered glycolytic enzymes, TPI and GAPDHS. PLoS ONE. 2013;8:e61434. doi: 10.1371/journal.pone.0061434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Okinaga S, Arai K. Metabolism of pachytene primary spermatocytes from rat testes: pyruvate maintenance of adenosine triphosphate level. Biol Reprod. 1984a;30:1187–1197. doi: 10.1095/biolreprod30.5.1187. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Okinaga S, Arai K. Metabolism of round spermatids: evidence that lactate is preferred substrate. Am J Physiol. 1984b;247:E234–242. doi: 10.1152/ajpendo.1984.247.2.E234. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Dai Q, Williams J, Goulding EH, Willis WD, Brown PR, Eddy EM. Disruption of a spermatogenic cell-specific mouse enolase 4 (Eno4) gene causes sperm structural defects and male infertility. Biol Reprod. 2013;88:90. doi: 10.1095/biolreprod.112.107128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien DA, Bellvé AR. Protein constituents of the mouse spermatozoon. I. An electrophoretic characterization. Dev Biol. 1980;75:386–404. doi: 10.1016/0012-1606(80)90171-2. [DOI] [PubMed] [Google Scholar]
- Odet F, Duan C, Willis WD, Goulding EH, Kung A, Eddy EM, Goldberg E. Expression of the gene for mouse lactate dehydrogenase C (Ldhc) is required for male fertility. Biol Reprod. 2008;79:26–34. doi: 10.1095/biolreprod.108.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romrell LJ, Bellvé AR, Fawcett DW. Separation of mouse spermatogenic cells by sedimentation velocity. A morphological characterization. Dev Biol. 1976;49:119–131. doi: 10.1016/0012-1606(76)90262-1. [DOI] [PubMed] [Google Scholar]
- Russell DL, Kim KH. Expression of triosephosphate isomerase transcripts in rat testis: Evidence for retinol regulation and a novel germ cell transcript. Biol Reprod. 1996;55:11–18. doi: 10.1095/biolreprod55.1.11. [DOI] [PubMed] [Google Scholar]
- Sakai I, Sharief FS, Li SS. Molecular cloning and nucleotide sequence of the cDNA for sperm-specific lactate dehydrogenase-C from mouse. Biochem J. 1987;242:619–622. doi: 10.1042/bj2420619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y, Tsai-Morris C-H, Dufau ML. Cell-specific and hormone-regulated expression of gonadotropin-regulated testicular RNA helicase gene (GRTH/Ddx25) resulting from alternative utilization of translation initiation codons in the rat testis. J Biol Chem. 2003;278:27796–27803. doi: 10.1074/jbc.M302411200. [DOI] [PubMed] [Google Scholar]
- Storey BT, Kayne FJ. Energy metabolism of spermatozoa. V. The Embden-Myerhof pathway of glycolysis: Activities of pathway enzymes in hypotonically treated rabbit epididymal spermatozoa. Fertil Steril. 1975;26:1257–1265. [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis AJ, Foster JA, Rosenbaum NA, Visconti PE, Gerton GL, Kopf GS, Moss SB. Targeting of a germ cell-specific type 1 hexokinase lacking a porin-binding domain to the mitochondria as well as to the head and fibrous sheath of murine spermatozoa. Mol Biol Cell. 1998;9:263–276. doi: 10.1091/mbc.9.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis AJ, Sui D, Riedel KD, Hofmann NR, Moss SB, Wilson JE, Kopf GS. A novel NH(2)-terminal, nonhydrophobic motif targets a male germ cell-specific hexokinase to the endoplasmic reticulum and plasma membrane. J Biol Chem. 1999;274:34467–34475. doi: 10.1074/jbc.274.48.34467. [DOI] [PubMed] [Google Scholar]
- Vemuganti SA, Bell TA, Scarlett CO, Parker CE, de Villena FP-M, O'Brien DA. Three male germline-specific aldolase A isozymes are generated by alternative splicing and retrotransposition. Dev Biol. 2007;309:18–31. doi: 10.1016/j.ydbio.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Vemuganti SA, Villena FP, de O'Brien DA. Frequent and recent retrotransposition of orthologous genes plays a role in the evolution of sperm glycolytic enzymes. BMC Genomics. 2010;11:285. doi: 10.1186/1471-2164-11-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visconti PE, Johnson LR, Oyaski M, Fornés M, Moss SB, Gerton GL, Kopf GS. Regulation, localization, and anchoring of protein kinase A subunits during mouse sperm capacitation. Dev Biol. 1997;192:351–363. doi: 10.1006/dbio.1997.8768. [DOI] [PubMed] [Google Scholar]
- Welch JE, Schatte EC, O'Brien DA, Eddy EM. Expression of a glyceraldehyde 3-phosphate dehydrogenase gene specific to mouse spermatogenic cells. Biol Reprod. 1992a;46:869–878. doi: 10.1095/biolreprod46.5.869. [DOI] [PubMed] [Google Scholar]
- Welch JE, Schatte EC, O'Brien DA, Eddy EM. Expression of a glyceraldehyde 3-phosphate dehydrogenase gene specific to mouse spermatogenic cells. Biol Reprod. 1992b;46:869–878. doi: 10.1095/biolreprod46.5.869. [DOI] [PubMed] [Google Scholar]
- Yuan PM, Dewan RN, Zaun M, Thompson RE, Gracy RW. Isolation and characterization of triosephosphate isomerase isozymes from human placenta. Arch Biochem Biophys. 1979;198:42–52. doi: 10.1016/0003-9861(79)90393-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
50 RACE of mouse mRNA. Utilization of 50 RACE identified the 50-UTR sequence of Tpi1 cDNAs from male mixed germ cells. Alignment of the four Tpi1 cDNA sequences: NM_009425.1 containing all three potential initiation codons, NM_009415.2 containing all three potential initiation codons, 50 RACE Transcript 1 containing all three potential initiation codons, and 50 RACE Transcript 2 containing only the second and third potential initiation codons.
30 RACE of mouse mRNA. Utilization of 30 RACE identified the 30-UTR sequence of Tpi1 cDNAs from male mixed germ cell populations. Alignment of the three Tpi1 cDNA sequences: NM_009425.1 is not predicted to contain a germ cell-specific 30-UTR, NM_009415.2 is predicted to contain a germ cell-specific 30-UTR, and the 30 RACE sequence. 30 RACE did not detect a predicted male germline-specific 30-UTR in mouse mixed germ cell cDNAs.
Amino acid sequences of TPI1 isoforms deduced from Tpi1 cDNA. Three putative translation initiation codons in full-length mouse Tpi1 cDNA (NM_009415.1) were discovered. The two orange boxes indicate putative translation initiation codons of the two male germ linespecific TPI1 isozymes, and the green box is the known translation initiation codon of somatic-type TPI1. The italicized numbers above the orange and green boxes indicate the amino acid lengths of the predicted proteins. The red box indicates the in-frame stop codon upstream from the start codons.


