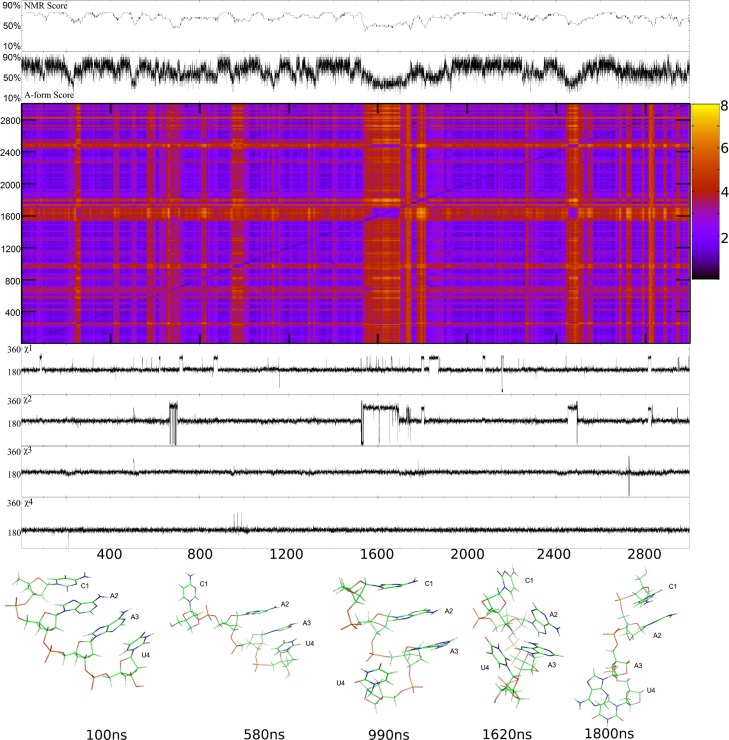
Figure 7.
L(CAAU) parm99_LNA simulation starting with the same A-form structure as the parm99 A-form simulation. The top two graphs are, respectively, the time dependence of NMR score averaged over 20 ns intervals and the A-form score in 0.1 ns intervals. The quilt plot is a 2D-RMS plot, where x and y axes are time, with blue and yellow areas representing, respectively, low and high RMSD (Å) between structures. The plot is perfectly symmetric along the diagonal. This allows visualization of structural clustering in the simulation. The bottom four plots represent χ torsions for nucleotides C1, A2, A3, and U4. All 2D-RMS plots were graphed with GNUPlot v 4.4. The mean NMR score is 66 ± 8%. The NMR distances for A3H1′-U4H1′ and A2H3′-A3H1′ are likely too short (Figure 5) due to spin diffusion, but were included in the scoring.