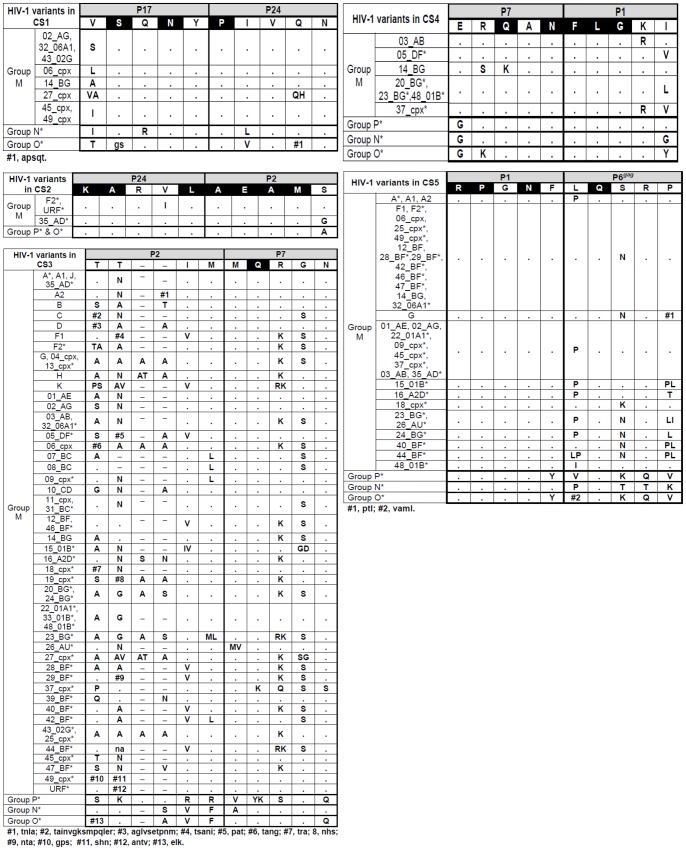
Figure 3. HIV-1 variants showing differences in CS1–CS5 amino acid vs. consensus-of-consensuses sequence from GenBank.
Changes are only indicated when they appeared in a specific position in at least 50% of the GenBank downloaded sequences in order to compare them with the GenBank consensus-of-consensuses sequence. Asterisks indicate the HIV-1 variants shown in Figure 2 with non available consensus sequence in GenBank. Black represents highly conserved amino acid residues and present in more than 99% of the 9,028 Gag and 3,906 GagPol HIV-1 sequences with respect to the consensus-of-consensuses sequence. When two residues within the analyzed sequences from each HIV-1 variant showed a conservation of 50% the two code letters were written in the upper case. When 3 or more residues appear in the same position and none presented a conservation of more than 50%, they were shown in lower case letters, which represented higher to lower conservation.