Abstract
Several studies have shown that synthesis of new proteins at the synapse is a prerequisite for the storage of long-term memories. Relatively little is known about the availability of distinct mRNA populations for translation at specific synapses, the process that determines mRNA localization, and the temporal designations of localized mRNA translation during memory storage. Techniques such as synaptosome preparation and microdissection of distal neuronal processes of cultured neurons and dendritic layers in brain slices are general approaches used to identify localized RNAs. Exploration of the association of RNA-binding proteins to the axonal transport machinery has led to the development of a strategy to identify RNAs that are transported from the cell body to synapses by molecular motor kinesin. In this article, RNA localization at the synapse, as well as its mechanisms and significance in understanding long-term memory storage, are discussed.
Keywords: RNA transport, RNAseq, kinesin, local translation, memory storage, signaling network, synapses
Groundbreaking observations by Steward and Levy on polysome localization at the base of dendritic spines challenged the view that proteins present at the synapse are synthesized in the cell body and transported to the synapses.1 Their observations suggested that mRNAs and the machinery for protein translation are transported to synapses. Several later studies have shown that RNAs are localized to distal parts of neurons in both vertebrates and invertebrates. Sequencing of RNAs prepared from microdissected neuronal processes of sensory neurons of Aplysia led to the identification of a few hundred RNAs that are enriched in neuronal processes.2,3 Using a microarray-based approach, RNAs localized to dendrites of hippocampal neurons were identified.4 Recently, RNaseq analysis identified a few thousand RNAs localized to the dendritic layer of the hippocampus.5
What Is the Significance of Transcriptome Localized to Synapses?
Several studies have shown that RNAs localized to synapses are used for synthesizing new proteins, which are necessary for synaptogenesis and activity-dependent synaptic remodeling. Local protein synthesis has a significant role in long-term memory storage (LTM) in the marine snail Aplysia,6-10 the fruit fly Drosophila,11,12 and in mice.13-16 Localization of specific mRNAs provides an efficient regulatory mechanism for restricting gene expression to specific subcellular locations in neurons and an elegant mechanism for synapse-specific plasticity.9,17 It allows individual synapses to undergo specific changes such as remodeling and growth in response to specific stimuli, such as learning. Such modifications can occur independently of unstimulated synapses in a persistent, protein synthesis-dependent manner. In a study using bifurcated sensory neurons of the marine snail Aplysia, Martin and colleagues described the role of mRNA translation during synapse-specific long-term facilitation (LTF).9 Synapse-specific translation is also critical for long-term potentiation (LTP) in the hippocampus.18 LTF and LTP are considered the cellular analogs of learning and memory storage. These studies utilized pharmacological inhibition of translation and electrophysiological measurements of the consequences of protein synthesis inhibition. Recently, synaptic translation of specific mRNAs has been visualized in the sensory neurons of Aplysia19 and in Drosophila20,21 by state-of-the-art imaging methodologies.
What Determines Localization and Composition of Transcriptome at the Synapse?
There are two main factors that determine localization of specific RNAs and composition of synaptic transcriptome: transcriptional activation of specific genes in the nucleus, and active transport of mRNAs from cell body to synapses. Studies that have used models such as Aplysia, Drosophila, and mice suggest that activation of several genes occurs during learning and memory processes. In the isolated Aplysia sensory to motor neuron cultures and in the intact animal, repeated exposure to serotonin (5-HT) causes a larger increase in cAMP, leading to the activation and translocation of PKA and MAP kinase to the nucleus. This translocation activates CREB1-dependent transcription and represses CREB2, leading to the induction of several immediate early genes.8,22-25 A similar sequence of second messenger signaling and gene induction was also found to have been recruited for long-term memory storage in Drosophila and in mice.26-31
Two specific genes of interest that are activated in Aplysia sensory neurons in response to 5-HT exposure are specific isoforms of molecular motor kinesin heavy chain (ApKHC1), and kinesin light chain (ApKLC2). Kinesin was first identified by Brady32 and Vale et al.,33 and is composed of two heavy chains (KHC) and two light chains (KLC). The super families of kinesin proteins (KIFs) are the molecular motors that transport cargos along microtubules. More than 40 KIFs have been identified in mammals.34,35 Kinesins were found to mediate the transport of RNAs and proteins from cell body to synapses.34 To understand the functions of the KIFs, several biochemical and genetic attempts were made to identify molecules carried by KIFs. This has led to the identification of several cargo proteins. For example, KIF17 binds to mLin-10 to transport the NMDA receptor in dendrites.36,37 Using the tail region of KIF5 as bait in affinity chromatography, Kanai et al. identified 42 proteins, including several known RNA-binding proteins that interact with kinesin, as well as few transported mRNAs (CAMKII α and Arc).38
Is the Kinesin-Mediated Transport of Proteins and RNAs Important for LTM?
In response to 5-HT, a modulatory transmitter released during behavioral sensitization, a specific isoform of the kinesin-heavy chain ApKHC1, is transcriptionally upregulated in both pre- and post-synaptic neurons of the Aplysia gill withdrawal reflex. We find that ApKHC1 knockdown in either the pre- or post-synaptic neurons blocked the establishment of LTF. However, it did not affect short-term facilitation (STF) or persistence of LTF, suggesting that during the early phase of memory storage, kinesin transports critical molecules that are later used for persistence of memory (Fig. 1). Indeed, several synaptic proteins required for synapse formation (e.g., neurexin, neuroligin, piccolo, and bassoon) were found in the kinesin complex isolated from the CNS, which are required for the establishment of LTF.39,40
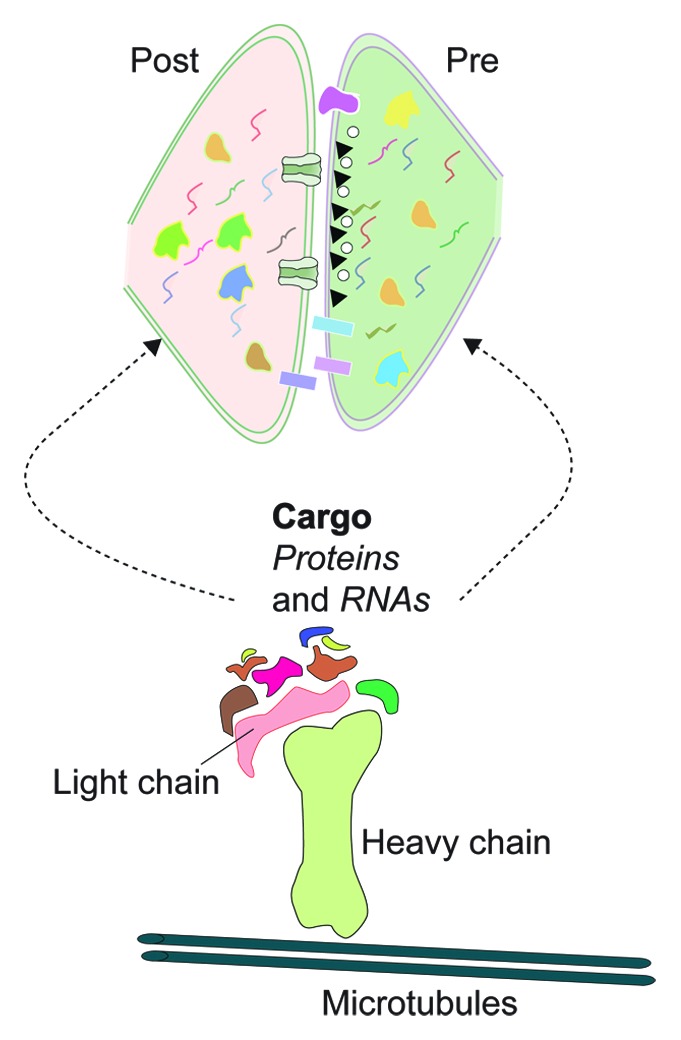
Figure 1. Kinesin-mediated transport of RNAs and protein regulate synaptic transcriptome and proteome. Molecular motor kinesin mediate transport of organelles, proteins and RNAs. Biochemical and genomic analysis of kinesin complexes from Aplysia has identified several proteins and RNAs as cargos transported to distal neuronal processes. Kinesin cargos regulate composition of transcriptome and proteome. Furthermore, kinesin cargos mediate several functions at the synapse such as formation of active zones, trans-synaptic signaling complexes, cytoskeletal re-arrangements, regulation of local translation, and signal transduction. Cartoon represents components of the transport machinery. Irregular shapes and squiggly lines represent protein and RNA cargos transported to pre and post-synaptic compartments.
Next, we searched for RNAs in the ApKHC1 complexes isolated from the Aplysia CNS. Since kinesin is the major motor that mediates the microtubule-dependent transport of gene products from the cell body to distal neuronal processes, we assumed that molecular characterization of the kinesin complex would identify RNAs transported to synapses. Furthermore, kinesin has been implicated in RNA transport in a variety of systems, from oocytes to neurons.43-47 Kinesin transports CaMKII α, Arc, and tau mRNAs in mammalian neurons,38,48,49 myelin basic protein mRNAs in oligodendrocytes,50 and oskar mRNA in Drosophila oocytes.51 Previous efforts have used cDNA library construction followed by Sanger sequencing2,3 and microarray studies to identify the composition of synaptic transcriptome.4,41,42 However, since the Sanger sequencing method is limited in the number of acquired reads and microarray studies that cannot identify new transcripts, a full repertoire of RNAs localized at synapses could not be characterized. We focused on the ApKHC1 transport complexes that contain several RNA-binding proteins such as staufen, FMRP, and CEPB, and NextGen sequencing methodologies (RNaseq) was used to identify all of the RNAs that are found in ApKHC1 complex.
The RNaseq characterization of ApKHC1 complex isolated from the CNS of Aplysia identified a few thousand mRNAs that were potentially transported to synapses.52 This unbiased analysis of RNAs in the ApKHC1 complex also identified several non-coding RNAs, such as antisense RNAs,52 whose specific function at the synapse is yet to be determined.
Our kinesin-based approach offers several advantages when compared with micro-dissection of dendritic regions: First, it allows the identification of RNAs based on their association with kinesin motors that transport gene products from cell body to synapses; second, RNAs associated with transport machinery reflects more dynamically regulated RNAs; third, RNAs that are actively transported in response to specific stimuli may be identified; and fourth, this methodology can easily be directly applied to identify synaptically targeted RNAs in regions of the CNS or neuronal cultures.
When and How Are the Synaptically Localized RNAs Utilized?
Understanding the temporal dynamics of translation of synaptic transcriptome and the mechanisms of their regulation of translation will help to identify specific signaling pathways and key mediators of synapse-specific memory storage. Based on the molecular, pharmacological, and electrophysiological data, it is apparent that the RNAs localized to synapses will be translated at different times during LTM storage. However, it is yet to be determined which RNAs are translated, as well as the determination of the temporal regulation of translation of RNAs during LTM storage. The newly translated proteins might be used to replenish proteins that are already present at the synapse specifically for the remodeling of the synapse and formation of new synapses. In order to obtain a comprehensive view of this process, the type of proteins and RNAs that are present at specific synapses and their dynamics during memory storage must be identified.
Miniaci et al.10 showed in Aplysia sensory-motor neuron synapses that inhibition of synaptic protein synthesis by local perfusion of emetine, or downregulation of ApCPEB (Aplysia cytoplasmic polyadenylation element binding protein) by local perfusion of specific antisense oligonucleotides against ApCPEB either at 24 or 48 h after 5-HT stimulation blocks LTF. These manipulations also cause selective retraction of newly formed sensory neuron varicosities induced by 5-HT. In contrast, the inhibition of local protein synthesis 72 h after 5-HT exposure has no effect on either synaptic growth or LTF. The CPEB proteins mediate the polyadenylation of RNAs and regulate local translation. These experiments demonstrated the differential requirement of translation during different phases of LTM storage.
Si et al.53,54 suggested that RNAs might be stored in the repressed form and utilized in response to specific synaptic stimulation. Si et al. also showed that ApCPEB is required, not for the initiation phase of LTF, but for the persistence phase of LTF. The LTF phenotype that is similar to CEPB knockdown was observed with a knockdown of eEF1 α, which is a regulator of translation. Injection of antisense oligonucleotides or antibodies that block the induction and expression of Ap-eEF1A did not affect initiation of LTF, but blocked its persistence.55 In a later study, we demonstrated that knockdown of ApKHC1 blocked the initiation of LTF; however, once the LTF is initiated, knockdown of ApKHC1 does not block persistence.40 Knockdown experiments of ApMHC, actin-dependent molecular motor, and mRNA cargo of ApKHC1, also showed similar results on initiation and persistence of LTF.52 Together, these studies suggest that during persistence of memory, synapses become autonomous with respect to transport from the cell body, and RNAs are stored at the synapses for later use.
CPEB is capable of activating these dormant mRNAs that contain CPE sequences by polyadenylation in response to specific stimulation.54,56-60 The P bodies are also described as storing dormant mRNAs.61,62 Another RNA-binding protein that is involved in translational regulation is Pumilio.63-67 An important aspect of this local mechanism is that translational repression can be removed at different times to meet the temporal requirements of these molecular substrates for the remodeling of existing synapses, and for the formation of new ones. A recent study demonstrated a RNA-based translational control of long-term memory storage. Rajasethupathy et al.68 showed that microRNA mir124 regulates the translation of several genes that are critical for LTF. More importantly the expression levels of mir124 are downregulated during LTF.
Is There a Molecular Code That Determines Synaptic Targeting of RNAs?
An obvious question that emerges from the identification of populations of RNAs that are localized to synapses is whether a molecular code ensures appropriate subcellular localization of RNAs. The 3′ or 5′ UTR sequences of RNAs might contain unique sequences that determine their subcellular localization. The existence of conserved sequence codes, which are similar to nuclear or mitochondrial or endoplasmic localization signals, are not as clear for synaptically localized RNAs. It appears that there is a distinct nucleotide sequence code specific to RNAs that determine its localization. A classic example is the “Zipcode” sequence present in actin mRNAs that are recognized by binding proteins described as “zipcode binding proteins” (ZBPs).69 Localization of β-actin mRNA to the leading edge of fibroblasts requires the presence of conserved elements in the 3′ untranslated region of the mRNA, including a 54-nucleotide element, termed “zipcode.” A protein of 68 kDa was identified, which binds to the proximal (to the coding region) half of the zipcode with high specificity (ZBP-1).70 Recently, Sladewski et al.71 showed the increasing number of localizing elements (zipcodes) on the mRNA-enhanced run length and event frequency of myosin V.
Like the binding of ZBP to β-actin mRNA, Vera, a homolog of ZBP, specifically binds to a repeated sequence motif in the Vg1 mRNA localization element required for its subcellular localization in Xenopus oocytes.72 Identification of specific short non-tandem repeat sequences that determine Vg1 mRNA localization in Xenopus oocytes leads to the development of a computational algorithm, REPFIND, which helps identify sequence repeats that seemingly determine the localization of RNAs in chordates.73,74 This analysis identified clusters of short CAC-containing motifs that determine the localization of virtually all mRNAs present in the vegetal cortex of Xenopus oocytes.74,75
What is the Significance of Transporting Several Thousand RNAs by Kinesin?
Bioinformatics analysis of RNAs transported by kinesin in the Aplysia central nervous system (CNS) has suggested that these RNAs are involved in signaling, translational control, cytoskeletal rearrangements, splicing, and metabolism. Large-scale analysis of transported RNAs provides new insights into sub-cellular regulation of signaling. For example, an important observation from this analysis is that RNAs of several neuropeptides are transported to synapses. Because neuropeptides need to be post-translationally processed before their release, identification of neuropeptide RNAs in the transport complex suggests that neuropeptide processing machinery must be present in the distal neuronal processes. Similarly, identification of RNAs involved in splicing machinery and rRNAs present at the synapses suggests the intriguing possibility that splicing and ribosome modification could occur in distal neuronal processes. Glanzer et al. have shown that splicing occurs in dendrites.79
Identifying RNAs that are transported and temporal regulation of their transport will help determine dynamic changes in the composition of the synaptic transcriptome. An obvious question that emerges from the large-scale analysis of RNAs is: “How do we determine specific roles of transported RNAs in memory storage?” We have begun to address this challenge by examining whether RNAs transported by kinesin are components of known signaling networks.80 The logic behind this analysis is that if specific RNAs form the nodes of signaling networks, such networks could be regulated by the availability of these RNAs, and by regulating the initiation of their translation. Once a specific network formed by transported RNAs is identified, specific nodes and pathways can be determined80 for molecular perturbation and functional experiments.
Conclusion
The process of addressing the aforementioned questions has begun to yield interesting insights into RNA localization, translational control, and memory storage. However, recent studies of RNA localization in mice,5 Aplysia,52 and proteomics analysis of signaling complexes at the synapse,76-78 suggest an unprecedented molecular complexity of the synapse. Translational control by miRNAs at the synapse adds another layer to this complexity. A caveat of large-scale studies using CNS or regions of the brain is that information about neuron-specific transport and localization is lost in the process. It is important to identify neuron-specific and circuit-specific synaptic transport of RNAs and proteins (Fig. 1), and study their temporal regulation. What is known so far about the dynamics and regulation of signaling at the synapse is clearly only the tip of the iceberg. Extensive and collaborative approaches are necessary so as to obtain deeper insight into signaling dynamics at synapses during memory storage.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
I sincerely thank the following organizations for funding support: the Whitehall Foundation, NIMH grant 1 R21MH096258-01A1, and The Scripps Research Institute, which enables me to carry out my research.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/27391
References
- 1.Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci. 1982;2:284–91. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moccia R, Chen D, Lyles V, Kapuya E, e Y, Kalachikov S, Spahn CM, Frank J, Kandel ER, Barad M, et al. An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J Neurosci. 2003;23:9409–17. doi: 10.1523/JNEUROSCI.23-28-09409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moroz LL, Edwards JR, Puthanveettil SV, Kohn AB, Ha T, Heyland A, Knudsen B, Sahni A, Yu F, Liu L, et al. Neuronal transcriptome of Aplysia: neuronal compartments and circuitry. Cell. 2006;127:1453–67. doi: 10.1016/j.cell.2006.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26:13390–9. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–66. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey CH, Kandel ER, Si K. The persistence of long-term memory: a molecular approach to self-sustaining changes in learning-induced synaptic growth. Neuron. 2004;44:49–57. doi: 10.1016/j.neuron.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 7.Casadio A, Martin KC, Giustetto M, Zhu H, Chen M, Bartsch D, Bailey CH, Kandel ER. A transient, neuron-wide form of CREB-mediated long-term facilitation can be stabilized at specific synapses by local protein synthesis. Cell. 1999;99:221–37. doi: 10.1016/S0092-8674(00)81653-0. [DOI] [PubMed] [Google Scholar]
- 8.Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–8. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 9.Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–38. doi: 10.1016/S0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- 10.Miniaci MC, Kim JH, Puthanveettil SV, Si K, Zhu H, Kandel ER, Bailey CH. Sustained CPEB-dependent local protein synthesis is required to stabilize synaptic growth for persistence of long-term facilitation in Aplysia. Neuron. 2008;59:1024–36. doi: 10.1016/j.neuron.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGuire SE, Deshazer M, Davis RL. Thirty years of olfactory learning and memory research in Drosophila melanogaster. Prog Neurobiol. 2005;76:328–47. doi: 10.1016/j.pneurobio.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Tully T, Preat T, Boynton SC, Del Vecchio M. Genetic dissection of consolidated memory in Drosophila. Cell. 1994;79:35–47. doi: 10.1016/0092-8674(94)90398-0. [DOI] [PubMed] [Google Scholar]
- 13.Frey U, Krug M, Reymann KG, Matthies H. Anisomycin, an inhibitor of protein synthesis, blocks late phases of LTP phenomena in the hippocampal CA1 region in vitro. Brain Res. 1988;452:57–65. doi: 10.1016/0006-8993(88)90008-X. [DOI] [PubMed] [Google Scholar]
- 14.Huang YY, Nguyen PV, Abel T, Kandel ER. Long-lasting forms of synaptic potentiation in the mammalian hippocampus. Learn Mem. 1996;3:74–85. doi: 10.1101/lm.3.2-3.74. [DOI] [PubMed] [Google Scholar]
- 15.Krug M, Lössner B, Ott T. Anisomycin blocks the late phase of long-term potentiation in the dentate gyrus of freely moving rats. Brain Res Bull. 1984;13:39–42. doi: 10.1016/0361-9230(84)90005-4. [DOI] [PubMed] [Google Scholar]
- 16.Stanton PK, Sarvey JM. Blockade of long-term potentiation in rat hippocampal CA1 region by inhibitors of protein synthesis. J Neurosci. 1984;4:3080–8. doi: 10.1523/JNEUROSCI.04-12-03080.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin KC, Zukin RS. RNA trafficking and local protein synthesis in dendrites: an overview. J Neurosci. 2006;26:7131–4. doi: 10.1523/JNEUROSCI.1801-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frey U, Morris RG. Synaptic tagging and long-term potentiation. Nature. 1997;385:533–6. doi: 10.1038/385533a0. [DOI] [PubMed] [Google Scholar]
- 19.Wang DO, Kim SM, Zhao Y, Hwang H, Miura SK, Sossin WS, Martin KC. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science. 2009;324:1536–40. doi: 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 21.Chen YC, Lin YQ, Banerjee S, Venken K, Li J, Ismat A, Chen K, Duraine L, Bellen HJ, Bhat MA. Drosophila neuroligin 2 is required presynaptically and postsynaptically for proper synaptic differentiation and synaptic transmission. J Neurosci. 2012;32:16018–30. doi: 10.1523/JNEUROSCI.1685-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alberini CM, Ghirardi M, Metz R, Kandel ER. C/EBP is an immediate-early gene required for the consolidation of long-term facilitation in Aplysia. Cell. 1994;76:1099–114. doi: 10.1016/0092-8674(94)90386-7. [DOI] [PubMed] [Google Scholar]
- 23.Bartsch D, Ghirardi M, Skehel PA, Karl KA, Herder SP, Chen M, Bailey CH, Kandel ER. Aplysia CREB2 represses long-term facilitation: relief of repression converts transient facilitation into long-term functional and structural change. Cell. 1995;83:979–92. doi: 10.1016/0092-8674(95)90213-9. [DOI] [PubMed] [Google Scholar]
- 24.Dash PK, Hochner B, Kandel ER. Injection of the cAMP-responsive element into the nucleus of Aplysia sensory neurons blocks long-term facilitation. Nature. 1990;345:718–21. doi: 10.1038/345718a0. [DOI] [PubMed] [Google Scholar]
- 25.Hegde AN, Inokuchi K, Pei W, Casadio A, Ghirardi M, Chain DG, Martin KC, Kandel ER, Schwartz JH. Ubiquitin C-terminal hydrolase is an immediate-early gene essential for long-term facilitation in Aplysia. Cell. 1997;89:115–26. doi: 10.1016/S0092-8674(00)80188-9. [DOI] [PubMed] [Google Scholar]
- 26.Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, Silva AJ. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 27.Byers D, Davis RL, Kiger JA., Jr. Defect in cyclic AMP phosphodiesterase due to the dunce mutation of learning in Drosophila melanogaster. Nature. 1981;289:79–81. doi: 10.1038/289079a0. [DOI] [PubMed] [Google Scholar]
- 28.Davis RL, Cherry J, Dauwalder B, Han PL, Skoulakis E. The cyclic AMP system and Drosophila learning. Mol Cell Biochem. 1995;149-150:271–8. doi: 10.1007/BF01076588. [DOI] [PubMed] [Google Scholar]
- 29.Davis RL, Dauwalder B. The Drosophila dunce locus: learning and memory genes in the fly. Trends Genet. 1991;7:224–9. doi: 10.1016/0168-9525(91)90369-2. [DOI] [PubMed] [Google Scholar]
- 30.Silva AJ, Kogan JH, Frankland PW, Kida S. CREB and memory. Annu Rev Neurosci. 1998;21:127–48. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- 31.Yin JC, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, Tully T. Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell. 1994;79:49–58. doi: 10.1016/0092-8674(94)90399-9. [DOI] [PubMed] [Google Scholar]
- 32.Brady ST. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature. 1985;317:73–5. doi: 10.1038/317073a0. [DOI] [PubMed] [Google Scholar]
- 33.Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/S0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–26. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 35.Miki H, Setou M, Kaneshiro K, Hirokawa N. All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci U S A. 2001;98:7004–11. doi: 10.1073/pnas.111145398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagawa T, Setou M, Seog D, Ogasawara K, Dohmae N, Takio K, Hirokawa N. A novel motor, KIF13A, transports mannose-6-phosphate receptor to plasma membrane through direct interaction with AP-1 complex. Cell. 2000;103:569–81. doi: 10.1016/S0092-8674(00)00161-6. [DOI] [PubMed] [Google Scholar]
- 37.Setou M, Nakagawa T, Seog DH, Hirokawa N. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288:1796–802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- 38.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–25. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 39.Choi YB, Li HL, Kassabov SR, Jin I, Puthanveettil SV, Karl KA, Lu Y, Kim JH, Bailey CH, Kandel ER. Neurexin-neuroligin transsynaptic interaction mediates learning-related synaptic remodeling and long-term facilitation in aplysia. Neuron. 2011;70:468–81. doi: 10.1016/j.neuron.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puthanveettil SV, Monje FJ, Miniaci MC, Choi YB, Karl KA, Khandros E, Gawinowicz MA, Sheetz MP, Kandel ER. A new component in synaptic plasticity: upregulation of kinesin in the neurons of the gill-withdrawal reflex. Cell. 2008;135:960–73. doi: 10.1016/j.cell.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki S, Ono N, Furusawa C, Kashiwagi A, Yomo T. Experimental optimization of probe length to increase the sequence specificity of high-density oligonucleotide microarrays. BMC Genomics. 2007;8:373. doi: 10.1186/1471-2164-8-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eberwine J, Belt B, Kacharmina JE, Miyashiro K. Analysis of subcellularly localized mRNAs using in situ hybridization, mRNA amplification, and expression profiling. Neurochem Res. 2002;27:1065–77. doi: 10.1023/A:1020956805307. [DOI] [PubMed] [Google Scholar]
- 43.Bullock SL. Translocation of mRNAs by molecular motors: think complex? Semin Cell Dev Biol. 2007;18:194–201. doi: 10.1016/j.semcdb.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 44.Hirokawa N. mRNA transport in dendrites: RNA granules, motors, and tracks. J Neurosci. 2006;26:7139–42. doi: 10.1523/JNEUROSCI.1821-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–90. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 46.Kiebler MA, DesGroseillers L. Molecular insights into mRNA transport and local translation in the mammalian nervous system. Neuron. 2000;25:19–28. doi: 10.1016/S0896-6273(00)80868-5. [DOI] [PubMed] [Google Scholar]
- 47.Tübing F, Vendra G, Mikl M, Macchi P, Thomas S, Kiebler MA. Dendritically localized transcripts are sorted into distinct ribonucleoprotein particles that display fast directional motility along dendrites of hippocampal neurons. J Neurosci. 2010;30:4160–70. doi: 10.1523/JNEUROSCI.3537-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aronov S, Aranda G, Behar L, Ginzburg I. Visualization of translated tau protein in the axons of neuronal P19 cells and characterization of tau RNP granules. J Cell Sci. 2002;115:3817–27. doi: 10.1242/jcs.00058. [DOI] [PubMed] [Google Scholar]
- 49.Ohashi S, Koike K, Omori A, Ichinose S, Ohara S, Kobayashi S, Sato TA, Anzai K. Identification of mRNA/protein (mRNP) complexes containing Puralpha, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J Biol Chem. 2002;277:37804–10. doi: 10.1074/jbc.M203608200. [DOI] [PubMed] [Google Scholar]
- 50.Carson JH, Worboys K, Ainger K, Barbarese E. Translocation of myelin basic protein mRNA in oligodendrocytes requires microtubules and kinesin. Cell Motil Cytoskeleton. 1997;38:318–28. doi: 10.1002/(SICI)1097-0169(1997)38:4<318::AID-CM2>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 51.Brendza RP, Serbus LR, Duffy JB, Saxton WM. A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science. 2000;289:2120–2. doi: 10.1126/science.289.5487.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puthanveettil SV, Antonov I, Kalachikov S, Rajasethupathy P, Choi YB, Kohn AB, Citarella M, Yu F, Karl KA, Kinet M, et al. A strategy to capture and characterize the synaptic transcriptome. Proc Natl Acad Sci U S A. 2013;110:7464–9. doi: 10.1073/pnas.1304422110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–35. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 54.Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, Kim JH, Zhu H, Kandel ER. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003;115:893–904. doi: 10.1016/S0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- 55.Giustetto M, Hegde AN, Si K, Casadio A, Inokuchi K, Pei W, Kandel ER, Schwartz JH. Axonal transport of eukaryotic translation elongation factor 1alpha mRNA couples transcription in the nucleus to long-term facilitation at the synapse. Proc Natl Acad Sci U S A. 2003;100:13680–5. doi: 10.1073/pnas.1835674100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huarte J, Stutz A, O’Connell ML, Gubler P, Belin D, Darrow AL, Strickland S, Vassalli JD. Transient translational silencing by reversible mRNA deadenylation. Cell. 1992;69:1021–30. doi: 10.1016/0092-8674(92)90620-R. [DOI] [PubMed] [Google Scholar]
- 57.Kindler S, Wang H, Richter D, Tiedge H. RNA transport and local control of translation. Annu Rev Cell Dev Biol. 2005;21:223–45. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richter JD. Translational control during early development. Bioessays. 1991;13:179–83. doi: 10.1002/bies.950130406. [DOI] [PubMed] [Google Scholar]
- 59.Stutz A, Conne B, Huarte J, Gubler P, Völkel V, Flandin P, Vassalli JD. Masking, unmasking, and regulated polyadenylation cooperate in the translational control of a dormant mRNA in mouse oocytes. Genes Dev. 1998;12:2535–48. doi: 10.1101/gad.12.16.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wells DG, Dong X, Quinlan EM, Huang YS, Bear MF, Richter JD, Fallon JR. A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J Neurosci. 2001;21:9541–8. doi: 10.1523/JNEUROSCI.21-24-09541.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Melemedjian OK, Mejia GL, Lepow TS, Zoph OK, Price TJ. Bidirectional regulation of P body formation mediated by eIF4F complex formation in sensory neurons. Neurosci Lett. 2013 doi: 10.1016/j.neulet.2013.09.048. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeitelhofer M, Macchi P, Dahm R. Perplexing bodies: The putative roles of P-bodies in neurons. RNA Biol. 2008;5:244–8. doi: 10.4161/rna.6948. [DOI] [PubMed] [Google Scholar]
- 63.Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13:286–96. doi: 10.1016/S0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 64.Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2006;103:4487–92. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Menon KP, Sanyal S, Habara Y, Sanchez R, Wharton RP, Ramaswami M, Zinn K. The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron. 2004;44:663–76. doi: 10.1016/j.neuron.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 66.Nakahata S, Kotani T, Mita K, Kawasaki T, Katsu Y, Nagahama Y, Yamashita M. Involvement of Xenopus Pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech Dev. 2003;120:865–80. doi: 10.1016/S0925-4773(03)00160-6. [DOI] [PubMed] [Google Scholar]
- 67.Ye B, Petritsch C, Clark IE, Gavis ER, Jan LY, Jan YN. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol. 2004;14:314–21. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- 68.Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil SV, Russo JJ, Sander C, Tuschl T, Kandel E. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63:803–17. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127:441–51. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–65. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sladewski TE, Bookwalter CS, Hong MS, Trybus KM. Single-molecule reconstitution of mRNA transport by a class V myosin. Nat Struct Mol Biol. 2013;20:952–7. doi: 10.1038/nsmb.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deshler JO, Highett MI, Abramson T, Schnapp BJ. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr Biol. 1998;8:489–96. doi: 10.1016/S0960-9822(98)70200-3. [DOI] [PubMed] [Google Scholar]
- 73.Andken BB, Lim I, Benson G, Vincent JJ, Ferenc MT, Heinrich B, Jarzylo LA, Man HY, Deshler JO. 3′-UTR SIRF: a database for identifying clusters of whort interspersed repeats in 3′ untranslated regions. BMC Bioinformatics. 2007;8:274. doi: 10.1186/1471-2105-8-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Betley JN, Frith MC, Graber JH, Choo S, Deshler JO. A ubiquitous and conserved signal for RNA localization in chordates. Curr Biol. 2002;12:1756–61. doi: 10.1016/S0960-9822(02)01220-4. [DOI] [PubMed] [Google Scholar]
- 75.Choo S, Heinrich B, Betley JN, Chen Z, Deshler JO. Evidence for common machinery utilized by the early and late RNA localization pathways in Xenopus oocytes. Dev Biol. 2005;278:103–17. doi: 10.1016/j.ydbio.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 76.Coba MP, Pocklington AJ, Collins MO, Kopanitsa MV, Uren RT, Swamy S, Croning MD, Choudhary JS, Grant SG. Neurotransmitters drive combinatorial multistate postsynaptic density networks. Sci Signal. 2009;2:ra19. doi: 10.1126/scisignal.2000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grant SG. A general basis for cognition in the evolution of synapse signaling complexes. Cold Spring Harb Symp Quant Biol. 2009;74:249–57. doi: 10.1101/sqb.2009.74.033. [DOI] [PubMed] [Google Scholar]
- 78.Ryan TJ, Grant SG. The origin and evolution of synapses. Nat Rev Neurosci. 2009;10:701–12. doi: 10.1038/nrn2717. [DOI] [PubMed] [Google Scholar]
- 79.Glanzer J, Miyashiro KY, Sul JY, Barrett L, Belt B, Haydon P, Eberwine J. RNA splicing capability of live neuronal dendrites. Proc Natl Acad Sci U S A. 2005;102:16859–64. doi: 10.1073/pnas.0503783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Papin JA, Hunter T, Palsson BO, Subramaniam S. Reconstruction of cellular signalling networks and analysis of their properties. Nat Rev Mol Cell Biol. 2005;6:99–111. doi: 10.1038/nrm1570. [DOI] [PubMed] [Google Scholar]


