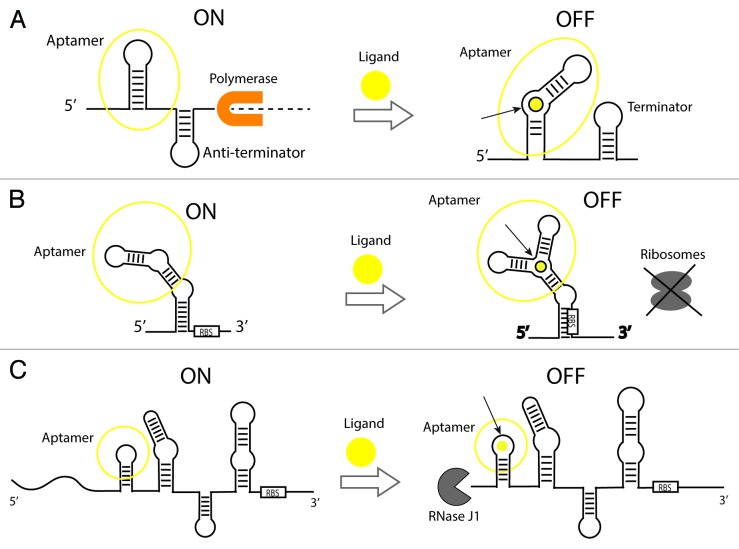
Figure 2. Gene expression control mechanisms of ligand-binding riboswitches. (A) Transcriptional elongation control by a ligand-activated riboswitch. In the absence of ligand, an anti-terminator stem is present allowing completion of transcription by a polymerase. A ligand binds a specific site in the aptamer and causes the formation of a terminator stem that arrests transcription and prevents the synthesis of a complete transcript (dashed line). The opposite mechanism in which transcription turns on upon binding has been identified in nature to a lesser extent (not shown in figure). (B) Translational initiation control by a ligand-activated riboswitch. In the presence of the ligand, a riboswitch undergoes a conformational change and sequesters the Ribosome-Binding Site (RBS) to prevent the ribosome from binding the mRNA. In the same way as with transcriptional controllers, there are examples of riboswitches that turn on translation upon binding (not shown in figure). (C) Translational control by an allosteric ribozyme. The glmS glucosamine-6-phosphate (GlcN6P)-activated ribozyme (aptazyme) is shown. In the absence of GlcN6P, translation is ON and the transcript is protected against degradation. In the presence of the ligand binding to the aptamer, the transcript is cleaved in the 5′ UTR by RNaseJ1 and is subject to decay.37

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.