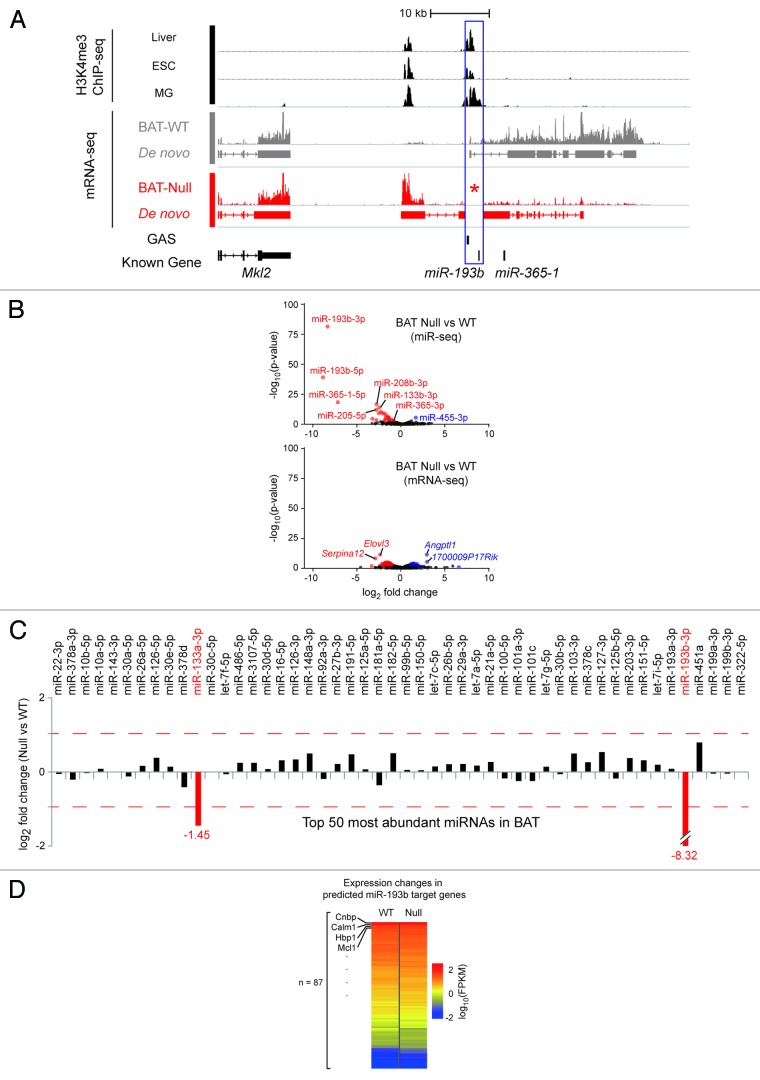
Figure 1. Features of the murine miR-193b-365-1 locus and molecular analyses of BAT in its absence. (A) The miR-193b-365-1 locus, located on chromosome 16, is positioned downstream of the Mkl2 gene and spans approximately 25 kbp. The structure of primary transcripts containing miR-193b and miR-365b sequences was determined using RNA-seq reads from interscapular BAT (BAT-WT) of two-day old mice. At least eight putative exons were identified that form several splice forms. Two distinct sets of H3K4me3 marks, obtained from publicly available ChIP-seq data sets in liver, embryonic stem cell (ESC) and mammary cells (MG) (blue box) indicate the presence of two distinct promoter/regulatory regions. None of the sequenced reads obtained from interscapular BAT from miR-193b−/− mice (BAT-Null) were mapped to this locus confirming complete deletion of miR-193b (asterisk). (B) Two different high-throughput sequencing approaches, mRNA-seq (whole transcriptome shotgun sequencing), and miR-seq (microRNA sequencing), revealed differentially expressed genes and microRNAs in the absence of miR-193b compared with wild-type. Blue and red dots indicate up- and downregulated genes (or miRs), respectively. (C) Expression ratios of top 50 highly abundant miRNAs (in descending order from left to right) between BAT-Null and BAT-WT are shown as log2-fold-change value. Red dashed lines indicate 2-fold changes. (D) A total of 87 genes were defined as miR-193b targets by at least four different algorithms via miRecords.30 Heatmap shows expression levels of the predicted miR-193b target genes. (E) qRT-PCR results from Ucp1 and Prdm16 (n = 3). Ucp1 in miR 193b-/-(CT 17.57 ± 0.35) and control BAT (16.34 ± 0.39); Prdm16 in miR 193b-/- (26.93 ± 0.02) and control BAT (26.71 ± 0.47).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.