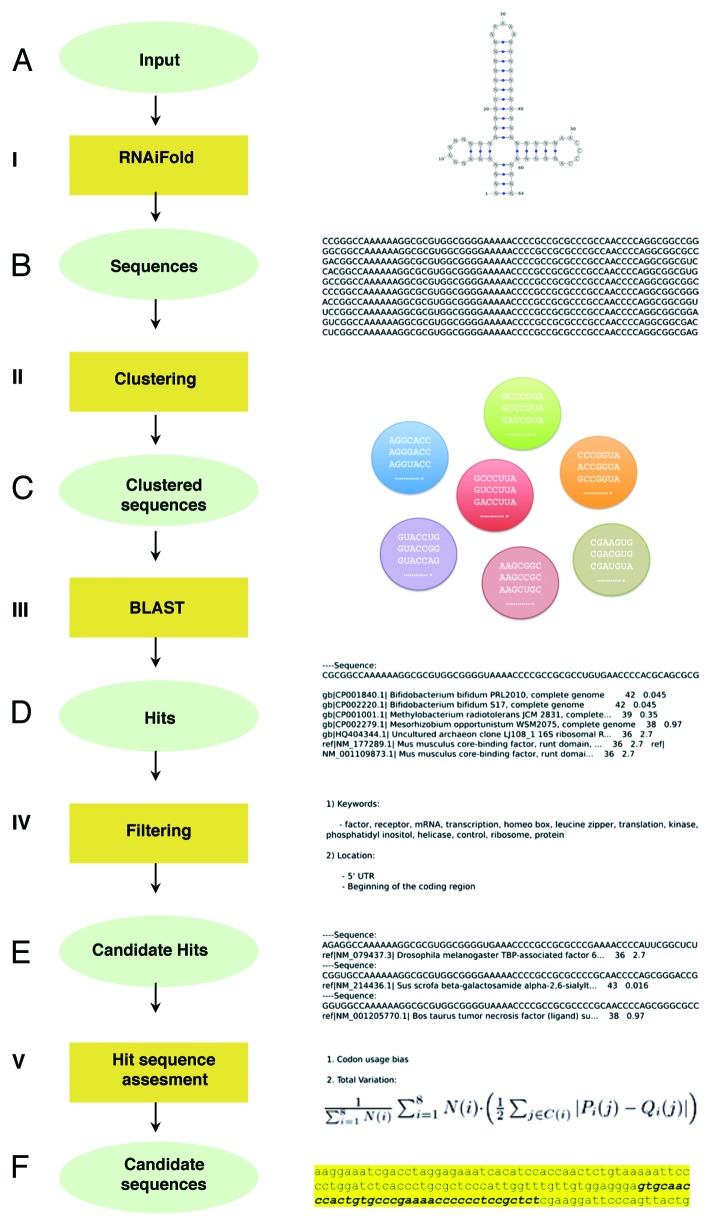
Figure 2. Computational pipeline summary. (A) RNA structure and sequence constraints input for RNAiFold (I). (B) Sequences returned that fold like the input structure were then clustered (II). (C) One representative of each cluster was BLASTed (III) with match/mismatch weights of 1/-1 and gap existence/extension penalties of 1/2. (D) Returned hits were filtered (IV) using relevant keywords and genomic locations. (E) Candidate hits located in coding sequences were assessed (V) by total variation and codon usage bias determination (using BitGene http://www.bitgene.com). (F) Candidate sequences selected for experimental validation.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.