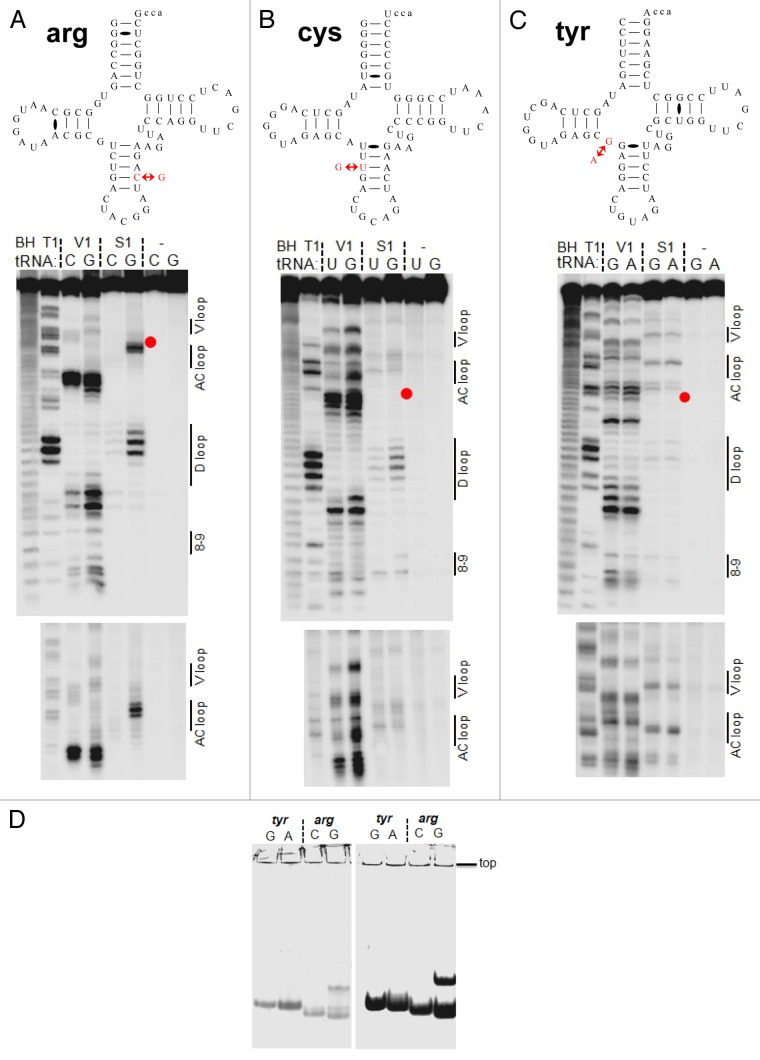
Figure 4. Structural mapping of pairs of tRNA isodecoder transcripts in vitro. Structural mapping of selected tRNA isodecoder pairs was performed by limited nuclease V1 and S1 digestion. Sequences in the context of standard tRNA secondary structure and mapping of tRNAArg(ACG) (A), tRNACys(GCA) (B), and tRNATyr(GUA) (C). Positions of RNA cuts were identified through alkaline hydrolysis (BH lane) and T1 ladder (T1 lane). The sequence change in the new tRNA isodecoder is indicated by red arrows and corresponds to the position of the red dot in gel. tRNA structural regions are indicated on the right side of the gel. (D) tRNA folding analysis by native gel electrophoresis. tRNA transcripts from T7 RNA polymerase reaction was directly loaded on 8% native gel containing 25 mM trisOAc, pH 7.5, 5 mM Mg(OAc)2. The amount of transcription reaction loaded in the left and right panels were 1 and 10 µl, respectively. Consistent with the structural mapping results, tRNATyr(GUA) variants fold in the same manner, whereas tRNAArg(ACG) variants fold very differently.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.