Abstract
In flaviviruses and hepatitis C virus (HCV), the NS3 gene encodes the N-terminal protease (NS3pro) and the C-terminal helicase (NS3hel). In HCV, the downstream NS4A is required for the NS3pro activity and exhibits a conserved EFDEMEE motif. To identify the role of this motif, we compared the ATPase and helicase activities of NS3 alone with those of the NS3-NS4A constructs. Our results suggest that the EFDEMEE motif is essential for regulating the ATPase activity of NS3hel. It is likely that this motif interferes with the ATP-binding site of NS3hel. It is becoming clear that NS4A functions as a cofactor of both proteinase and helicase in HCV.
HCV is a major cause of chronic hepatitis, which frequently results in liver cirrhosis and hepatocellular carcinoma in humans. HCV is a member of the genus Hepacivirus, family Flaviviridae, and is a relative of dengue virus (DV), West Nile virus (WNV), yellow fever virus, Japanese encephalitis virus and related flaviviruses. In the course of infection, the single-stranded, positive-strand, genomic RNA of HCV is translated into a polyprotein precursor. The precursor consists of the three structural proteins (C, E1 and E2) and the seven non-structural (NS) proteins arranged in the order NH2-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH [1]. The structural proteins (but not NS proteins) are components of mature virus particles. The precursor is inserted into the endoplasmic reticulum membrane and processed by host-cell and viral proteinases. The C-E1-E2-p7 junctions are processed by host cell proteinases. The NS2-NS3-NS4A-NS4B-NS5A-NS5B region is processed by the combined action of the viral cysteine (NS2-3) and serine (NS3) proteinases [1].
The NS2-3 proteinase catalyzes the cleavage between NS2 and NS3. The NS3 proteinase (NS3pro) represents the N-end, ~180-residue, domain of the 631-residue NS3 protein. The C-end domain of NS3 encodes the ATP-dependent RNA helicase (NS3hel) [2]. In HCV, NS4A is required for full activity of NS3pro [3]. This cofactor function of HCV NS4A is similar to that of NS2B of flaviviruses, which is a cofactor of NS3pro in DV and WNV [4, 5]. The NS2B cofactor is localized upstream of NS3 in the polyprotein precursor [6]. As a result, the regulation of the proteinase and helicase activities in WNV and DV involves both the upstream NS2B and the downstream NS4A, which are the cofactors of NS3pro and NS3hel, respectively [4, 5, 7]. It is established that in HCV the NS4A cofactor binds directly to the N-terminal portion of NS3pro [8]. As a result, it is likely that the NS3pro and NS3hel domains of NS3 are more interdependent in HCV than in DV or WNV [7, 9] and that the single NS4A performs a dual function in regulating the NS3pro-NS3hel activities in HCV.
We have demonstrated that the 1–53 N-terminal hydrophilic, cytoplasmic sequence of NS4A regulates the ATP-ase activity of NS3hel in WNV [10]. Consistently, Beran et al. [11] demonstrated that HCV NS4A promoted RNA-coupled ATP hydrolysis by NS3hel and that this effect was mediated by unspecified residues in the C-terminal acidic domain of NS4A. In agreement with this, the C-terminal acidic motif EELPD/E is essential for regulating the ATPase activity of WNV NS3hel [10]. This motif is conserved in both the flaviviruses and HCV (EFDEMEE in HCV). We hypothesized that the NS4A acidic motif is directly involved in the regulation of the ATPase activity of the HCV helicase. Our results support this hypothesis.
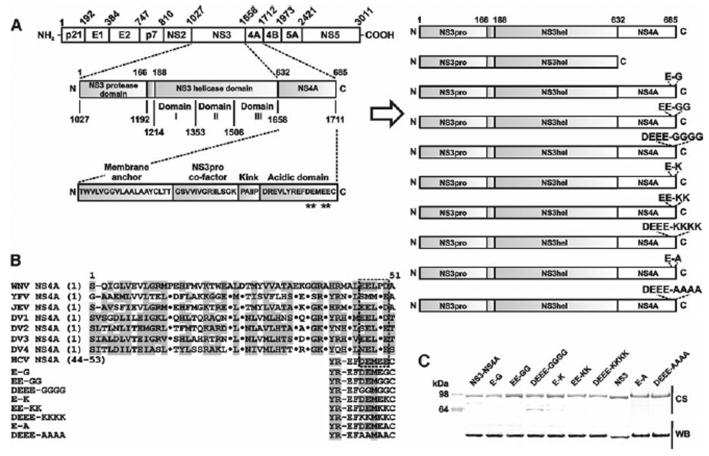
The HCV constructs were amplified by PCR using the HCV NS3-NS4A construct fused with the SUMO protein (provided by Kevin Raney, Univ. of Arkansas for Medical Sciences) [12] (Table 1). The NS3-NS4A sequence was derived from the HCV Con 1b sequence [13, 14]. The constructs were cloned in the pET101/D-TOPO vector (Invitrogen). We also constructed mutants in which the conserved Glu and Asp of the Asp1706-Glu-Met-Glu-Glu-Cys1711 motif of NS4A were replaced with either Gly or Ala and Lys. As a result, we obtained NS3-NS4A mutants (Fig. 1A, B). To facilitate protein purification, the constructs were N-terminally tagged with a 6xHis tag followed by SUMO. In the absence of the SUMO portion, the constructs precipitated, thus making any enzymatic analysis impractical.
Table 1.
Primers used for the cloning and mutagenesis of HCV constructs
| Construct | Primers |
|
|---|---|---|
| Forward | Reverse | |
| NS3-NS4A | 5′-CACCATGGGTCATCACCATCATCATCACGGGTCGGAC-3′ | 5′-TTAGCACTCTTCCATCTCATCGAACTCCCG-3′ |
| NS3 | 5′-TTAGCTCGTGACGACCTCCAGG-3′ | |
| NS3-NS4A E-G | 5′-TTAGCACCCTTCCATCTCATCGAACTCCCG-3′ | |
| NS3-NS4A EE-GG | 5′-TTAGCACCCCCCCATCTCATCGAACTCCCG-3′ | |
| NS3-NS4A DEEE-GGGG | 5′-TTAGCACCCCCCCATCCCCCCGAACTCCCG-3′ | |
| NS3-NS4A E-K | 5′-TTAGCACTTTTCCATCTCATCGAACTCCCG-3′ | |
| NS3-NS4A EE-KK | 5′-TTAGCACTTCTTCATCTCATCGAACTCCCG-3′ | |
| NS3-NS4A DEEE-KKKK | 5′-TTAGCACTTCTTCATCTTTTTGAACTCCCG-3′ | |
| NS3-NS4A E-A | 5′-TTAGCACGCTTCCATCTCATCGAACTCCCG-3′ | |
| NS3-NS4A DEEE-AAAA | 5′-TTAGCACGCTGCCATCGCAGCGAACTCCCG-3′ | |
Mutant positions are underlined
Fig. 1.
Mutant constructs. A, The structure of the HCV polyprotein precursor (left) and the recombinant NS3hel–NS4A constructs (right). All constructs included the NS3pro domain. The constructs were also N-terminally tagged with a 6xHis tag followed by a SUMO tag. Mutant positions in the acidic C-terminal domain of NS4A are indicated by asterisks. B, Sequence alignment of the flaviviral NS4A. WNV, West Nile virus; YFV, yellow fever virus; JEV, Japanese encephalitis virus; and DV1–4, dengue virus types 1–4. The acidic C-terminal motif is boxed. The 44–53 acidic motif of HCV NS4A is shown below the alignment. Homologous residues are shaded in grey. Dots indicate identical residues. C, SDS-gel electrophoresis of the purified recombinant proteins. CS, Coomassie staining; WB, western blotting with a 6xHis antibody
To express the NS3-NS4A constructs, E.coli BL21 CodonPlus (DE3)-RIPL cells (Stratagene) were transformed with the individual recombinant pET101/D-TOPO vectors. Transformed cells were grown in LB broth at 37°C to reach A600 = 0.6. Protein expression was induced for 16 h at 30°C using 1 mM IPTG. The collected cells were resuspended in 40 ml 20 mM Tris–HCl, pH 8.0, supplemented with 1 M NaCl, a proteinase inhibitor cocktail (Roche) and 1 mg/ml lysozyme, and disrupted by sonication. Cell debris was removed by centrifugation. The NS3-NS4A and mutant proteins were purified from the supernatant using a HiTrap Co2+- chelating Sepharose FastFlow column (GE Healthcare) and a 0–500 mM imidazole gradient. These procedures yielded the purified proteins, although minor impurities were present in the DEEE-GGGG samples (Fig. 1C). Our tests using the fluorescent peptide cleavage substrates suggested that the NS3pro remained catalytically inactive in the purified samples. As a result, the NS4A portion remained covalently linked with the NS3hel domain. Several independent lots of the purified proteins were isolated and tested.
The ATPase activity of the constructs was measured multiple times in triplicate in wells of a 96-well plate using the NTPase Assay colorimetric system (Innova Biosciences). The unwinding activity of the NS3hel enzyme requires the presence of ATP (5 mM) in the in vitro reactions [7, 11]. In turn, the ATPase activity does not require the binding of NS3hel to a nucleic acid or polynucleotide substrate. Therefore, the reactions did not include the oligonucleotide substrates. In addition to the NTPase assay components, the reactions (100 μl each) contained 20 mM Tris–HCl, pH 7.5, and the purified protein samples (100 nM). The resulting absorbance was measured at A620 on a fluorescence reader. The rate of ATP hydrolysis was quantified using a standard curve.
Double-stranded DNA substrates with a 20-nt single-stranded terminus were formed by annealing an 18-bp sequence (5′-GCCTCGCTGCCGTCGCCA-3′; D1) to a 38-bp oligonucleotide (5′-TGGCGACGGCAGCGAGGC TTTTTTTTTTTTTTTTTTTT-3′; D2). D1 (50 pmol) was 5′-labelled using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]-ATP (Perkin Elmer). The labeled product was separated from the free label using a Micro Bio-Spin 6 column (Bio-Rad). To generate duplex DNA, labeled D1 (50 pmol) was mixed with D2 (100 pmol) in 20 mM Tris–HCl, pH 7.8, containing 150 mM NaCl and 0.1 mM EDTA. The samples were boiled for 1 min and then cooled to 20°C. Duplex formation was confirmed by non-denaturing 10% acrylamide gel electrophoresis followed by radioautography.
The unwinding activity assay was performed as described earlier [7]. The unwinding activity of the constructs was measured multiple times in 200-μl reactions containing 20 mM Tris–HCl, pH 7.8, supplemented with 25 mM NaCl, 3 mM MgCl2, 2 mM DTT, 20 mg BSA, 5 mM ATP, 500 nM unlabeled D1 oligonucleotide (as a trap oligo), 5 nM DNA oligonucleotide preformed duplex and protein sample (200 nM). Aliquots (20 μl) were withdrawn from the reactions at 5–30 min. The reactions were stopped by adding 6 μl 50 mM EDTA. Single-stranded oligonucleo-tides were separated from the duplex by non-denaturing 10% acrylamide gel electrophoresis followed by autoradiography. Gel images were digitized using Multi Gauge software (Fujifilm).
To test if the acidic EFDEMEE motif that is localized in the C-terminal portion of NS4A is involved in the regulation of the ATPase activity of HCV NS3pro, we measured the unwinding and ATPase activities of the NS3 protein alone and also those of the wild-type and mutant HCV NS3-NS4A constructs. For this purpose, we constructed a soluble HCV NS3-NS4A chimera that included the full-length NS3 protein and the individual NS3 1027–1657 protein lacking the NS4A sequence. In the NS3-NS4A constructs, the full-length, 53-residue, 1658–1711 NS4A sequence, which included the membrane anchor portion, was directly linked to the 1027–1657 NS3pro-hel sequence of the HCV polyprotein precursor. To facilitate the isolation and analysis of the recombinant proteins from E. coli cells, the constructs were tagged with a Hisx6 tag. We also constructed and tested mutants in which the conserved negatively charged Glu and Asp of the Asp1706-Glu-Met-Glu-Glu-Cys1711 motif of NS4A were replaced with either Gly or Ala and Lys. Based on the amino acid substitutions they contained, these mutant were called E–G, E-A and E-K, EE-GG and EE-KK, DEEE-GGGG, DEEE-AAAA and DEEE-KKKK (Fig. 1A, B). In our experimental conditions and because of the presence of the protease inhibitor cocktail in the samples, we did not detect any significant autoproteolysis of the purified NS3-NS4A proteins—a fortunate event that is in contrast with the earlier observations by others [11]. The immunoreactivity with the His-tag antibody implied that the N-terminal His-SUMO tag was retained in the purified constructs.
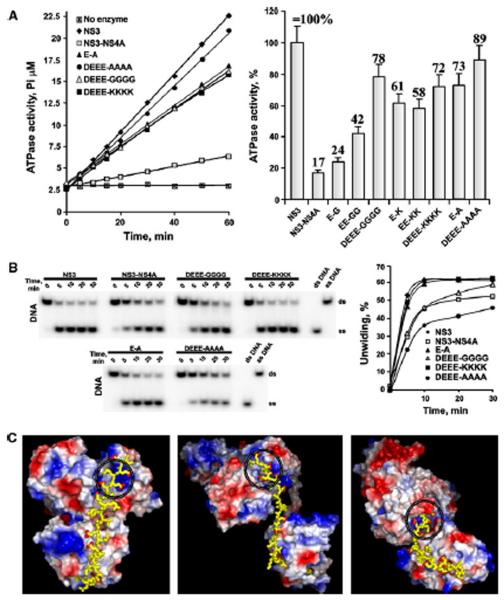
A direct comparison of the NS3 and NS3-NS4A constructs demonstrated that NS3-NS4A had an approximately 6-fold reduced ATPase activity compared to NS3 alone (Fig. 2A). Further analysis of the NS4A mutants showed that the DEEE-GGGG, DEEE-AAAA and DEEE-KKKK mutations restored the ATPase activity of the NS3-NS4A constructs most efficiently, implying that less ATP might be consumed from a same amount of unwinding activity [15] and that by decreasing the rate of ATP hydrolysis and simultaneously either increasing or keeping the unwinding activity constant, protein–protein interactions play an important role in the viral replication complex. In general, the Ala and Lys residues caused a more pronounced effect then the Gly substitutions. Obviously, the acidic EFDEMEE motif plays a role in regulating the ATPase activity of the HCV helicase. These data support and extend the findings of others [10, 11, 15].
Fig. 2.
NS4A mutants affect the ATPase activity of NS3 helicase. A, The ATPase activity of the wild-type and mutant constructs. Left, the reactions were performed for the indicated times using 1 mM ATP. Right, the ATPase activity of the constructs (at the 30 min time point) relative to NS3 alone (set as 100%). Error bars indicate SEM. Experiments were repeated multiple times with similar results. The E-G, EE-GG, E-K and EE-KK mutants are omitted in panel B for clarity. B, NS4A mutants do not significantly affect the unwinding activity of NS3 helicase. Left, the DNA unwinding efficiency of the mutants. Right, the images on the left were digitized, and the band density was calculated. The time course of the conversion of the ds-DNA substrate band into the ss-DNA product band is shown to demonstrate the initial rate of DNA duplex unwinding. Experiments were repeated multiple times with similar results. The E-G, EE-GG, E-K and EE-KK mutants are omitted for the clarity. ds and ss, double-and single-stranded, respectively. C, Three alternative potential structures of HCV NS3-NS4A. The structures are shown as surface models of NS3hel and NS3pro [the top and the bottom domains, respectively, colored according to their electrostatic potential (blue, positive; red, negative)]. The C-terminal sequence AIIPDREVLYREFDEMEEC of HCV NS4A is shown as yellow sticks. Red dots indicate the carboxyl oxygens of Asp and Glu. The ATP-binding site is circled. The models were build using PyMol, with the crystal structures of HCV (PDB 1CU1; left), Murray Valley encephalitis virus (PDB 2WV9; middle) and DV4 NS3 (PDB 2VBC; right) as templates
The NS3 protein by itself demonstrated a high unwinding activity (Fig. 2B). The wild-type NS3-NS4A construct was less efficient. The activity of the DEEE–GGGG and DEEE-AAAA mutants was similar to that of the wild-type NS3-NS4A, while the activity of the E-A, DEEE-KKKK and other mutants was more comparable to that of NS3 alone. These findings suggest that NS4A did not significantly affect the initial rate at which the NS3hel constructs unwound the oligonucleotide duplex. It appears that NS4A allowed the NS3-NS4A constructs to accomplish the substrate unwinding while consuming lower quantities of ATP compared to NS3 alone. Our results directly complement recent observations by others [11] who determined that HCV NS4A enhanced the ability of NS3hel to bind RNA in the presence of ATP, thereby acting as a cofactor for helicase activity in HCV. Overall, it is now becoming increasingly clear that in HCV and other members of the family Flaviviridae, the acidic motif of NS4A functions as an NS3hel cofactor.
Because the N-terminal part tethers NS4A to the membrane, only the hydrophilic, 34-residue C-terminal portion of NS4A is flexible and available for interactions with NS3pro and NS3hel. The model we build is not intended to predict the precise structure of the NS3-NS4A complex but only to estimate if the length of the hydrophilic portion is sufficient for the interactions of the C-end acidic motif of NS4A with the ATP-binding site in NS3hel. The crystal structures of HCV NS3 with the 14-residue NS4A fragment (PDB 1CU1) [8] and of DV4 NS3 (PDB 2VBC) [16] and Murray Valley encephalitis virus NS3 (PDB 2WV9) [17] in the complex with the NS2B cofactor were used as templates to model the HCV NS3-NS4A complex. In 1CU1, the C-terminal portion of NS4A (AAIIPDREVLYREFDEMEEC) was added directly as an extended β-strand to the known structure of the N-end, 14-residue portion of NS4A. In the models using 2VBC and 2WV9, the N-end portion of NS4A was placed proximal to the NS3pro active site while the C-terminal portion was placed into the NS3hel structure. Regardless of the distinct relative conformations of the NS3pro and NS3hel domains, these models suggest that the length of the 34-residue hydrophilic portion of NS4A is sufficient for its interactions with both NS3pro and NS3hel in the HCV NS3 protein. It appears possible that the C-terminal EFDEMEE motif of NS4A is close to the positively-charged ATP-binding site of the NS3hel domain, while the N-terminal portion of NS4A is proximal to the NS3pro active site (Fig. 2C).
Our data imply that NS4A functions as a cofactor of both NS3pro and NS3hel of HCV NS3 and provide a biochemical rationale for an additional role of NS4A in replication of viruses of the family Flaviviridae [18]. As a result, NS4A may be considered a novel and promising drug target in HCV and also in the flaviviruses.
Acknowledgments
This study was supported by NIH grants RR020843 and AI055789 (AYS).
Footnotes
Publisher's Disclaimer: Your article is protected by copyright and all rights are held exclusively by Springer-Verlag. This e-offprint is for personal use only and shall not be self-archived in electronic repositories. If you wish to self-archive your work, please use the accepted author's version for posting to your own website or your institution's repository. You may further deposit the accepted author's version on a funder's repository at a funder's request, provided it is not made publicly available until 12 months after publication.
References
- 1.Dubuisson J. Hepatitis C virus proteins. World J Gastroenterol. 2007;13:2406–2415. doi: 10.3748/wjg.v13.i17.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki R, Suzuki T, Ishii K, Matsuura Y, Miyamura T. Processing and functions of Hepatitis C virus proteins. Intervirology. 1999;42:145–152. doi: 10.1159/000024973. [DOI] [PubMed] [Google Scholar]
- 4.Aleshin AE, Shiryaev SA, Strongin AY, Liddington RC. Structural evidence for regulation and specificity of flaviviral pro-teases and evolution of the Flaviviridae fold. Protein Sci. 2007;16:795–806. doi: 10.1110/ps.072753207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erbel P, Schiering N, D'Arcy A, Renatus M, Kroemer M, Lim SP, Yin Z, Keller TH, Vasudevan SG, Hommel U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol. 2006;13:372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- 6.Sampath A, Padmanabhan R. Molecular targets for flavivirus drug discovery. Antiviral Res. 2009;81:6–15. doi: 10.1016/j.antiviral.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chernov AV, Shiryaev SA, Aleshin AE, Ratnikov BI, Smith JW, Liddington RC, Strongin AY. The two-component NS2B-NS3 proteinase represses DNA unwinding activity of the West Nile virus NS3 helicase. J Biol Chem. 2008;283:17270–17278. doi: 10.1074/jbc.M801719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao N, Reichert P, Taremi SS, Prosise WW, Weber PC. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease–helicase. Structure. 1999;7:1353–1363. doi: 10.1016/s0969-2126(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 9.Beran RK, Pyle AM. Hepatitis C viral NS3-4A protease activity is enhanced by the NS3 helicase. J Biol Chem. 2008;283:29929–29937. doi: 10.1074/jbc.M804065200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiryaev SA, Chernov AV, Aleshin AE, Shiryaeva TN, Strongin AY. NS4A regulates the ATPase activity of the NS3 helicase: a novel cofactor role of the non-structural protein NS4A from West Nile virus. J Gen Virol. 2009;90:2081–2085. doi: 10.1099/vir.0.012864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beran RK, Lindenbach BD, Pyle AM. The NS4A protein of hepatitis C virus promotes RNA-coupled ATP hydrolysis by the NS3 helicase. J Virol. 2009;83:3268–3275. doi: 10.1128/JVI.01849-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sikora B, Chen Y, Lichti CF, Harrison MK, Jennings TA, Tang Y, Tackett AJ, Jordan JB, Sakon J, Cameron CE, Raney KD. Hepatitis C virus NS3 helicase forms oligomeric structures that exhibit optimal DNA unwinding activity in vitro. J Biol Chem. 2008;283:11516–11525. doi: 10.1074/jbc.M708125200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tackett AJ, Chen Y, Cameron CE, Raney KD. Multiple full-length NS3 molecules are required for optimal unwinding of oligonucleotide DNA in vitro. J Biol Chem. 2005;280:10797–10806. doi: 10.1074/jbc.M407971200. [DOI] [PubMed] [Google Scholar]
- 14.Mackintosh SG, Lu JZ, Jordan JB, Harrison MK, Sikora B, Sharma SD, Cameron CE, Raney KD, Sakon J. Structural and biological identification of residues on the surface of NS3 helicase required for optimal replication of the hepatitis C virus. J Biol Chem. 2006;281:3528–3535. doi: 10.1074/jbc.M512100200. [DOI] [PubMed] [Google Scholar]
- 15.Xu T, Sampath A, Chao A, Wen D, Nanao M, Chene P, Vasudevan SG, Lescar J. Structure of the Dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 A. J Virol. 2005;79:10278–10288. doi: 10.1128/JVI.79.16.10278-10288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo D, Xu T, Hunke C, Gruber G, Vasudevan SG, Lescar J. Crystal structure of the NS3 protease–helicase from dengue virus. J Virol. 2008;82:173–183. doi: 10.1128/JVI.01788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Assenberg R, Mastrangelo E, Walter TS, Verma A, Milani M, Owens RJ, Stuart DI, Grimes JM, Mancini EJ. Crystal structure of a novel conformational state of the flavivirus NS3 protein: implications for polyprotein processing and viral replication. J Virol. 2009;83:12895–12906. doi: 10.1128/JVI.00942-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roosendaal J, Westaway EG, Khromykh A, Mackenzie JM. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J Virol. 2006;80:4623–4632. doi: 10.1128/JVI.80.9.4623-4632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]