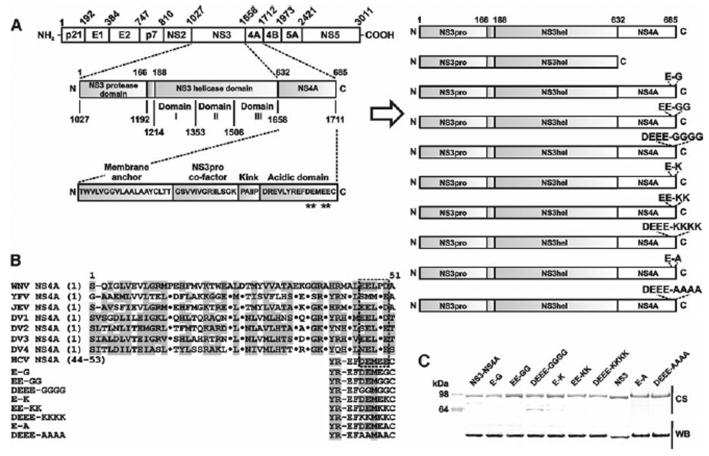
Fig. 1.
Mutant constructs. A, The structure of the HCV polyprotein precursor (left) and the recombinant NS3hel–NS4A constructs (right). All constructs included the NS3pro domain. The constructs were also N-terminally tagged with a 6xHis tag followed by a SUMO tag. Mutant positions in the acidic C-terminal domain of NS4A are indicated by asterisks. B, Sequence alignment of the flaviviral NS4A. WNV, West Nile virus; YFV, yellow fever virus; JEV, Japanese encephalitis virus; and DV1–4, dengue virus types 1–4. The acidic C-terminal motif is boxed. The 44–53 acidic motif of HCV NS4A is shown below the alignment. Homologous residues are shaded in grey. Dots indicate identical residues. C, SDS-gel electrophoresis of the purified recombinant proteins. CS, Coomassie staining; WB, western blotting with a 6xHis antibody