Abstract
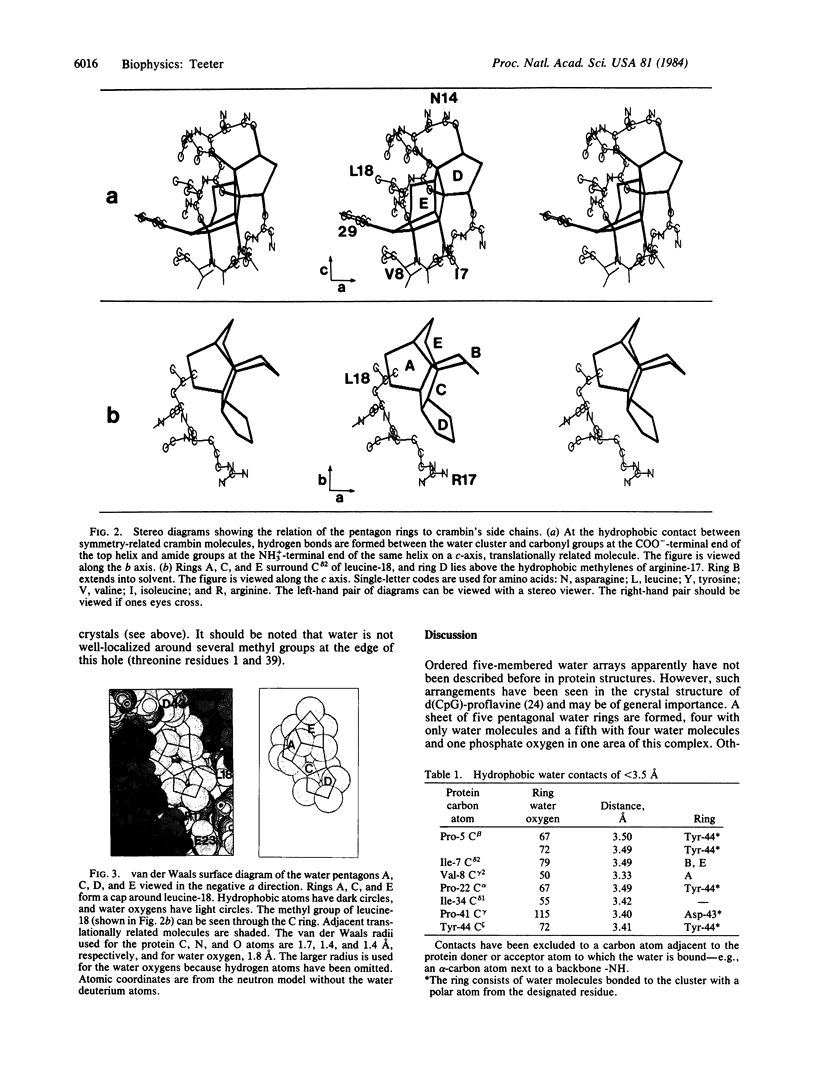
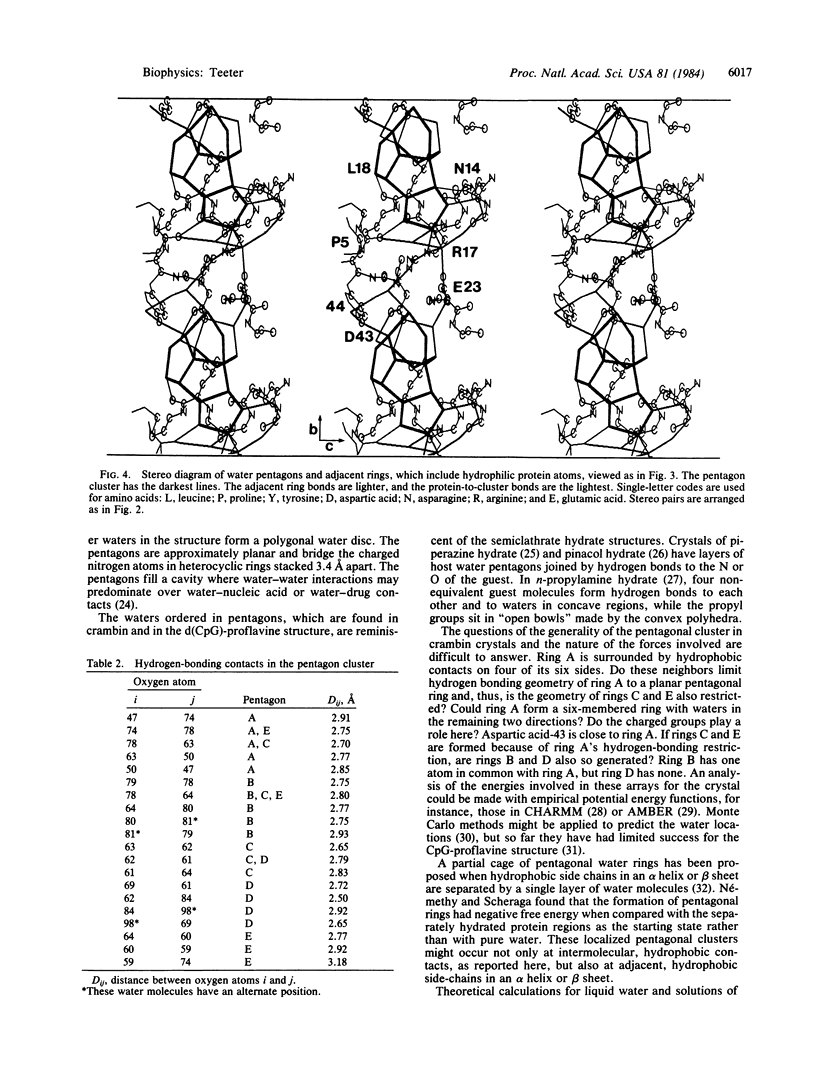
The water structure has been analyzed for a model of the protein crambin refined against 0.945-Å x-ray diffraction data. Crystals contain 32% solvent by volume, and 77% of the solvent molecules have been located—i.e., 2 ethanol molecules and 64 water molecules with 10-14 alternate positions. Many water oxygen atoms found form chains between polar groups on the surface of the protein. However, a cluster of pentagonal arrays made up of 16 water molecules sits at a hydrophobic, intermolecular cleft and forms a cap around the methyl group of leucine-18. Several waters in the cluster are hydrogen-bonded directly to the protein. Additional closed circular arrays, which include both protein atoms and other water oxygen atoms, form next to the central cluster. This water array stretches in the b lattice direction between groups of three ionic side chains.
Keywords: x-ray diffraction, clathrate hydrate, liquid-like
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blake C. C., Pulford W. C., Artymiuk P. J. X-ray studies of water in crystals of lysozyme. J Mol Biol. 1983 Jul 5;167(3):693–723. doi: 10.1016/s0022-2836(83)80105-3. [DOI] [PubMed] [Google Scholar]
- Carter C. W., Jr, Kraut J., Freer S. T., Nguyen-Huu-Xuong, Alden R. A., Bartsch R. G. Two-Angstrom crystal structure of oxidized Chromatium high potential iron protein. J Biol Chem. 1974 Jul 10;249(13):4212–4225. [PubMed] [Google Scholar]
- Edsall J. T., McKenzie H. A. Water and proteins. II. The location and dynamics of water in protein systems and its relation to their stability and properties. Adv Biophys. 1983;16:53–183. doi: 10.1016/0065-227x(83)90008-4. [DOI] [PubMed] [Google Scholar]
- Eklund H., Nordström B., Zeppezauer E., Söderlund G., Ohlsson I., Boiwe T., Brändén C. I. The structure of horse liver alcohol dehydrogenase. FEBS Lett. 1974 Aug 25;44(2):200–204. doi: 10.1016/0014-5793(74)80725-8. [DOI] [PubMed] [Google Scholar]
- James M. N., Sielecki A. R. Structure and refinement of penicillopepsin at 1.8 A resolution. J Mol Biol. 1983 Jan 15;163(2):299–361. doi: 10.1016/0022-2836(83)90008-6. [DOI] [PubMed] [Google Scholar]
- KAUZMANN W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- KLOTZ I. M. Protein hydration and behavior; many aspects of protein behavior can be interpreted in terms of frozen water of hydration. Science. 1958 Oct 10;128(3328):815–822. doi: 10.1126/science.128.3328.815. [DOI] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Madison V., Osguthorpe D. J., Dauber P., Hagler A. T. Monte Carlo simulations of peptide solvation. Biopolymers. 1983 Jan;22(1):27–31. doi: 10.1002/bip.360220106. [DOI] [PubMed] [Google Scholar]
- Mezei M., Beveridge D. L., Berman H. M., Goodfellow J. M., Finney J. L., Neidle S. Monte Carlo studies on water in the dCpG/proflavin crystal hydrate. J Biomol Struct Dyn. 1983 Oct;1(1):287–297. doi: 10.1080/07391102.1983.10507440. [DOI] [PubMed] [Google Scholar]
- Neidle S., Berman H. M., Shieh H. S. Highly structured water network in crystals of a deoxydinucleoside---drug complex. Nature. 1980 Nov 13;288(5787):129–133. doi: 10.1038/288129a0. [DOI] [PubMed] [Google Scholar]
- Rees D. C., Lewis M., Lipscomb W. N. Refined crystal structure of carboxypeptidase A at 1.54 A resolution. J Mol Biol. 1983 Aug 5;168(2):367–387. doi: 10.1016/s0022-2836(83)80024-2. [DOI] [PubMed] [Google Scholar]
- Rose G. D. Prediction of chain turns in globular proteins on a hydrophobic basis. Nature. 1978 Apr 13;272(5654):586–590. doi: 10.1038/272586a0. [DOI] [PubMed] [Google Scholar]
- Teeter M. M., Hendrickson W. A. Highly ordered crystals of the plant seed protein crambin. J Mol Biol. 1979 Jan 15;127(2):219–223. doi: 10.1016/0022-2836(79)90242-0. [DOI] [PubMed] [Google Scholar]
- Watenpaugh K. D., Margulis T. N., Sieker L. C., Jensen L. H. Water structure in a protein crystal: rubredoxin at 1.2 A resolution. J Mol Biol. 1978 Jun 25;122(2):175–190. doi: 10.1016/0022-2836(78)90034-7. [DOI] [PubMed] [Google Scholar]






