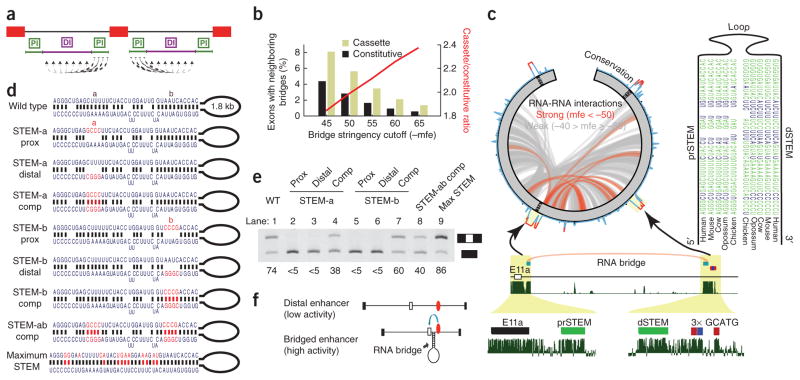
Figure 5.
An RNA bridge between ENAH E11a and a conserved distal RBFOX site is necessary for exon inclusion. (a) A schematic of the strategy used to find RNA bridges. Regions proximal to exons were tested for the ability to pair to all positions in the distal region in the same intron. (b) The fraction of cassette (light green) and constitutive (black) exons with neighboring predicted RNA bridges as the negative minimum free energy (mfe) threshold for defining an RNA bridge is made more stringent. The ratio between these fractions is depicted as a red line. (c) All predicted RNA-RNA interactions within E11-E11a-E12 (green) of the ENAH pre-mRNA are displayed in a circos plot (at left). Paired regions are classified as strong (mfe ≤ −50 kcal per mol; red) or weak (mfe > −50 kcal per mol; gray). PhastCons conservation scores are illustrated on the circumference of the circle. A stem-loop structure that is conserved across mammalian and even avian genomes is shown with base-paired nucleotides indicated in green. The location of this RNA bridge is shown in detail below. Vertebrate conservation from phastCons is represented as continuous density in dark green. (d) Wild-type and mutated (red letters) RNA duplex structures are shown. (e) RT-PCR analysis of ENAH structural mutants in transfected T47D cells. Labels above each numbered lane correspond to experiments using each of the structures in d. Ψ is listed below each lane. Prox, proximal; comp, compensatory mutations. (f) Model illustrating the function of the RNA bridge to position Rbfox sites (red ovals) close to an exon to regulate splicing.