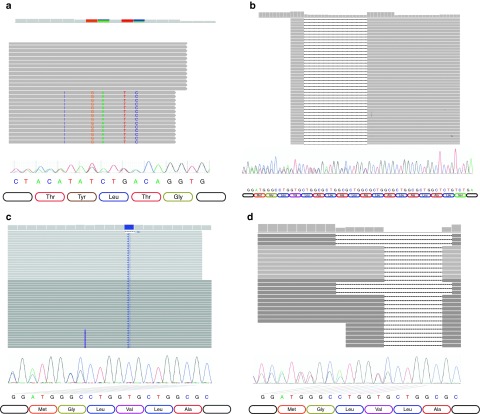
Figure 3.
Genotyping by Assembly-Templated Alignment correctly genotypes insertions and deletions that are undetectable by the alignment-only method. Read from top to bottom, each panel provides tracks for cumulative depth of coverage (vertical gray bars); representative MIP alignments (horizontal gray bars) with mismatches (colored letters), insertions (purple bars), and gaps (dashed lines); chromatogram; reference DNA and amino acid sequence for (a) heterozygous BLM c.2207_2212delinsTAGATTC in sample GM04408 as well as several alleles in the first exon of SMPD1 including (b) a heterozygous 18-bp deletion in sample GM20342 (minus strand), (c) a heterozygous 12-bp insertion and homozygous substitution in sample GM17282 (plus strand), and (d) compound heterozygous 6- and 12-bp deletions in sample GM00502 (minus strand). Chromatogram trace offsets corresponding to specific heterozygous insertion and deletion patterns are indicated with slanted lines color coded by reference base. For clarity, offsets are shown for (c) and (d) only. MIP, molecular inversion probe.