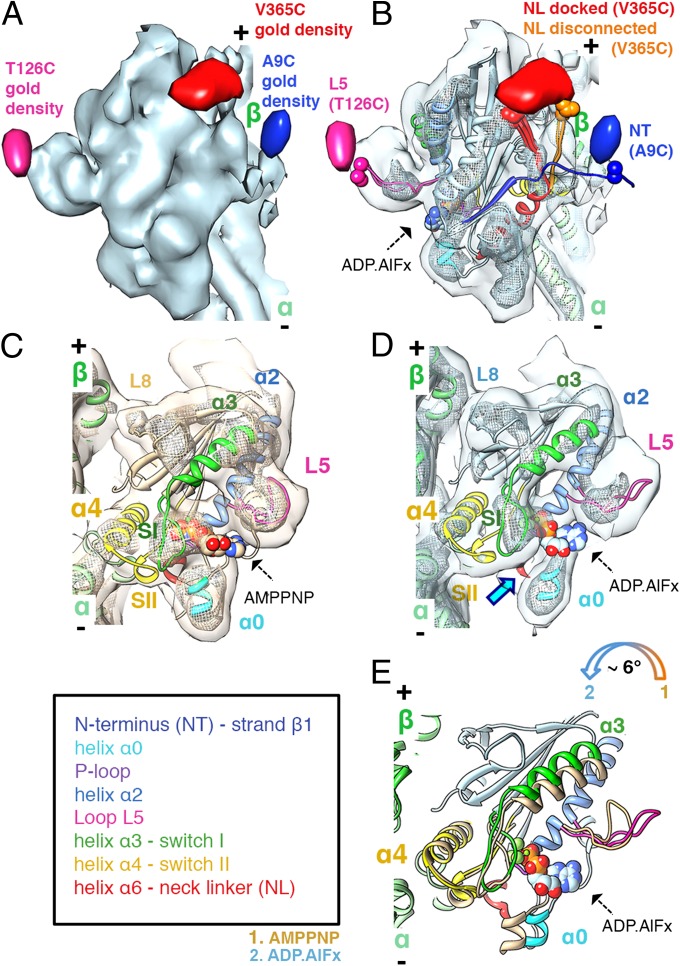
Fig. 1.
The ATP hydrolysis transition state: MD tilting, L5 uncurling, and CNB destabilization. (A) MT-bound K5 MD in the ADP.AlFx state (light blue surface: 1.1σ contour) with gold difference maps superimposed (blue: 2.3σ; pink: 2.2σ; red: 2.2σ contour). In all figures, the MT plus end is towards the top. (B) Pseudo-atomic models of K5 MD (color-coded as in the key) bound to an αβ-tubulin dimer (green ribbon) docked into the reconstruction (contour as in A; mesh: 3.3σ contour, ∼1/3 of the volume). Residues to which gold clusters were covalently attached and the nucleotide analogue are shown in space-filling representation. NL conformers clustered into docked (red) and disconnected (orange) conformations are indicated (SI Materials and Methods). (C and D) View towards the nucleotide-binding pocket of the AMPPNP (15) (C: coral surface: 1σ contour, mesh: 3σ contour, to depict the same molecular volumes as in B) and ADP.AlFx (D: contoured as in B). The blue arrow indicates the absence of contact between helix α0 and switch I in the ADP.AlFx state, which is present in the AMPPNP state. (E) Comparison of K5 MD models of ADP.AlFx (colored as in the key) and AMPPNP states [coral (15)], superimposed on helices α4. ATP hydrolysis triggers a tilt of the MD towards the MT lattice (colored arrow). L5 uncurls from its arch-like conformation upon ATP hydrolysis (comparison based on superimposed helices α2). Displacements of helix α3/switch I and helix α0 are caused by MD tilting.