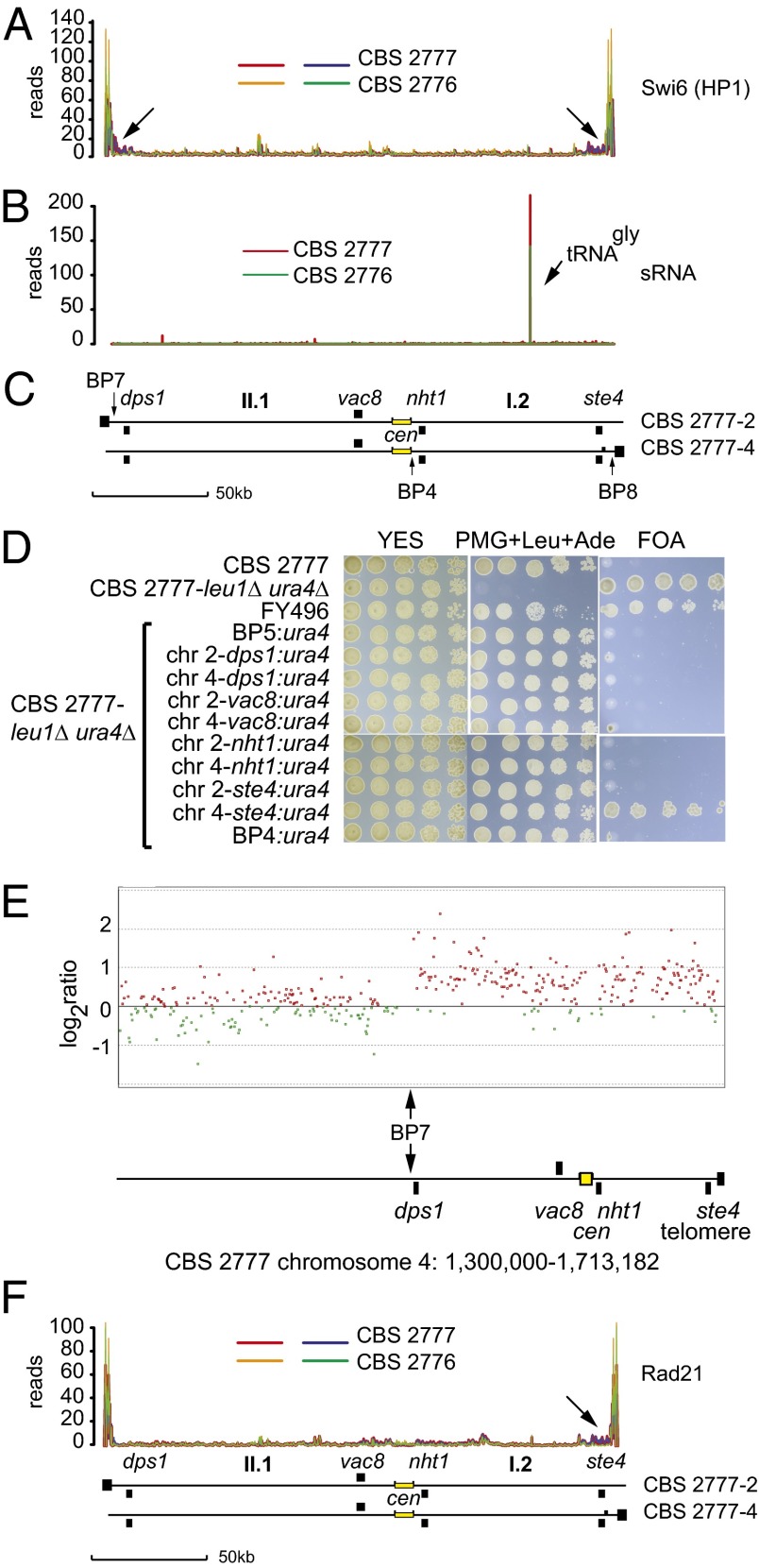
Fig. 4.
Absence of heterochromatin or an increase in Rad21 binding around the centromeres of CBS 2777 chromosomes 2 and 4. (A) Comparison of the binding of Swi6 (HP1) to the pericentric DNA held in common by CBS 2777 chromosomes 2 and 4 in strains CBS 2777 and CBS 2776 as measured by ChIP-seq on GFP epitope-tagged Swi6 (HP1). The data from the duplicate experiments of the CBS 2777 are shown in red and blue, and those from CBS 2776 are shown in green and orange. Arrows indicate the increased binding of Swi6 to dps1 and ste4 in CBS 2777. (B) Small RNAs derived from the pericentric DNA held in common by CBS 2777 chromosomes 2 and 4 in strains CBS 2777 and CBS 2776. (C) Organization of the pericentric DNA held in common by CBS 2777 chromosomes 2 and 4 aligned with respect to the data shown in A and B. (D) Absence of gene silencing in the pericentric DNA of CBS 2777 chromosomes 2 and 4. Silencing activity was assayed by testing the growth of URA+ cells on 5-FOA. Cells were grown to saturation in YES and inoculated in serial fivefold dilutions starting from an initial concentration of ∼7.5 × 103 on agar plates of the indicated composition. In FY496 (27), the ura4 gene was placed into the imr1L repeat of 972h−; this strain served as a positive control for the silencing seen in CBS 2777. (E) Absence of repression of gene expression around the acrocentric centromeres of CBS 2777 chromosome 4. Total RNA was extracted from either CBS 2777 or CBS 2776 cells, and cRNA probes were prepared and labeled with CY3 or CY5, respectively, and analyzed by competitive hybridization to a custom 4 × 44K gene expression microarray. The results were processed and analyzed using Agilent Genespring software. Red and green points mark genes with greater or lesser expression, respectively, in CBS 2777 compared with CBS 2776. The region around the centromere of CBS 2777 chromosome 4 is illustrated. The vertical arrow marks the junction with the region duplicated on chromosome 2 defined by breakpoint 7. (F) Comparison of the binding of Rad21 to the pericentric DNA held in common by CBS 2777 chromosomes 2 and 4 in strains CBS 2777 and CBS 2776 as measured by ChIP-seq on PK9 epitope-tagged Rad21. The arrow indicates the increase in binding of Rad21 to ste4 in CBS 2777. The traces in F are aligned with those in A and B, and this is indicated by the idiogramatic representation (C) of the centromeric region of chromosomes 2 and 4 placed below the three sets of traces.