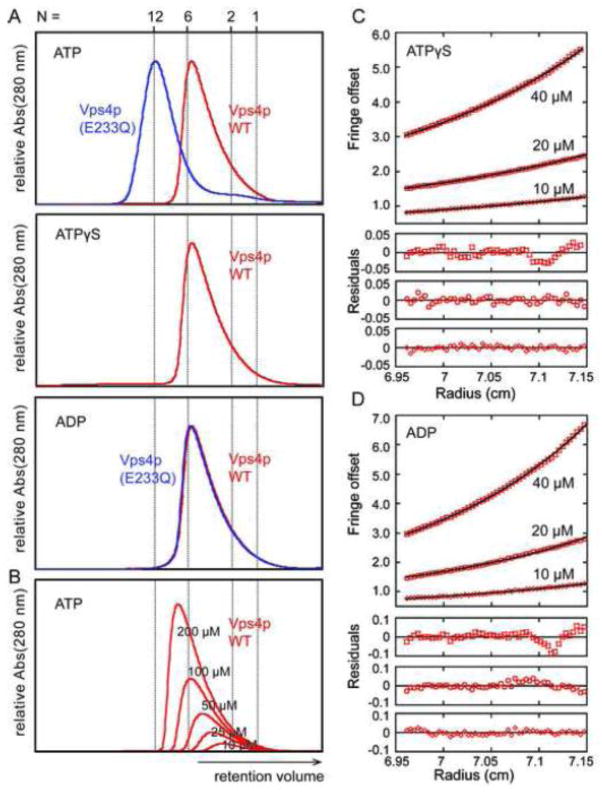
Figure 1.
Oligomerization of S. cerevisiae Vps4p proteins. (A) Size exclusion chromatograms of wild-type Vps4p (red) and Vps4p(E233Q) (blue), injected at a concentration of 100 μM, in the presence of 2 mM magnesium chloride and 1 mM ATP, 0.2 mM ATPγS, or 1 mM ADP. Expected retention volumes for different oligomeric states of Vps4p based on molecular weight standards are indicated with dotted lines. Note that Vps4p(E233Q) elutes as a dodecamer in the presence of ATP, but as a hexamer in the presence of ADP, whereas wild-type Vps4p elutes as a hexamer in the presence of ATP, ATPγS or ADP. (B) Size exclusion chromatograms of wild-type Vps4p at concentrations ranging from 10 μM to 200 μM in the presence of 1 mM ATP. Note that the complex migrates more slowly at lower concentrations, indicating that Vps4p is in a rapid equilibrium between different oligomeric states. (C, D) Equilibrium analytical ultracentrifugation at 4 °C indicates that wild-type Vps4p is in a dimer-hexamer equilibrium in the presence of (C) 1 mM ATPγS or (D) 1 mM ADP. The interference signal from sedimentation data at 5000 rpm is plotted versus the distance from the axis of rotation (radius) as red symbols. Three different protein concentrations are displayed: 40 μM (upper), 20 μM (middle) and 10 μM (lower). The global fit to data obtained for three concentrations at rotor speeds of 3000 rpm (not shown) and 5000 rpm using a dimer-hexamer model is shown in black with residuals for all three concentrations displayed below.