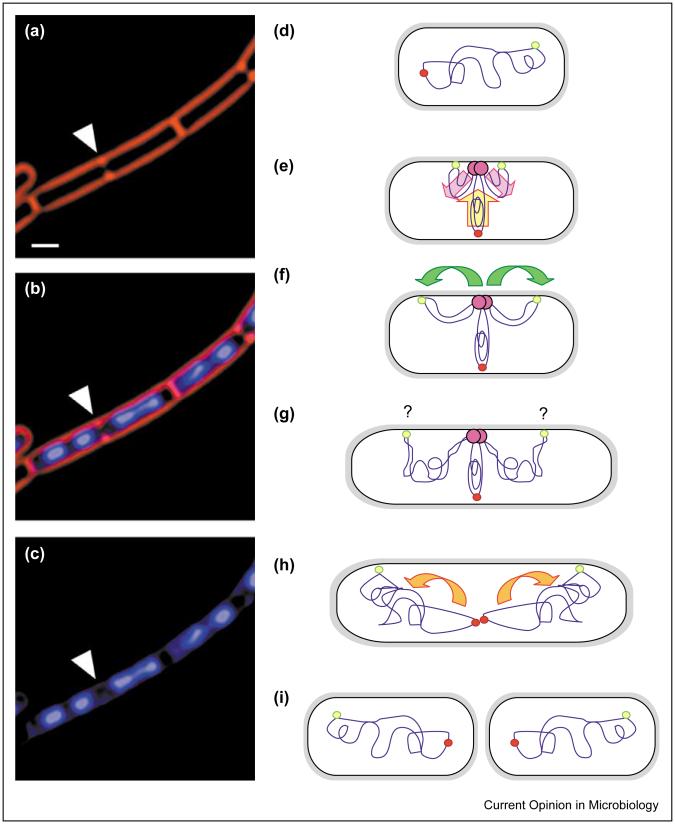
Figure 1.
Bacterial chromosome segregation. (a-c) Micrograph showing rapidly growing B. subtilis cells stained with FM 4-64 to visualize cell membranes (red) and DAPI to show chromosomal DNA (blue). During rapid growth, B. subtilis cells remain in septate chains because cell separation lags behind division, and multiple chromosomal replication cycles occur to enable rapid division. Thus, in cells that are about to divide (arrowhead), chromosome segregation often appears complete in the nascent daughter cells. In this case, the nascent daughter cell on the left has two apparently separate chromosomes, whereas that on the right has a bi-lobed nucleoid, which is likely to represent a partially replicated or segregated chromosome. Scale bar = 1 μm. (d-i) Model for chromosome segregation in slowly growing E. coli and B. subtilis. (d) In newborn cells, the origin (yellow-green circle) and terminus (red circle) appear at opposite cell poles. (e) At the initiation of chromosome replication, the chromosome rotates to bring the origin to the replisome (pink circle). The replicated DNA is extruded towards opposite cell poles (pink arrows), whereas the unreplicated DNA moves towards the replisome (yellow arrow). (f) The replicated origin DNA moves towards the cell poles (green arrows), (g) where it is anchored by an unidentified origin-binding protein (purple question mark). (h) The daughter chromosomes are separately condensed during replication by the concerted action of SMC and topoisomerases (orange arrows). (i) Finally, division results in two polarized daughter cells, in which the termini lie at each new pole, whereas the origins are at each older pole.