Fig. 4.
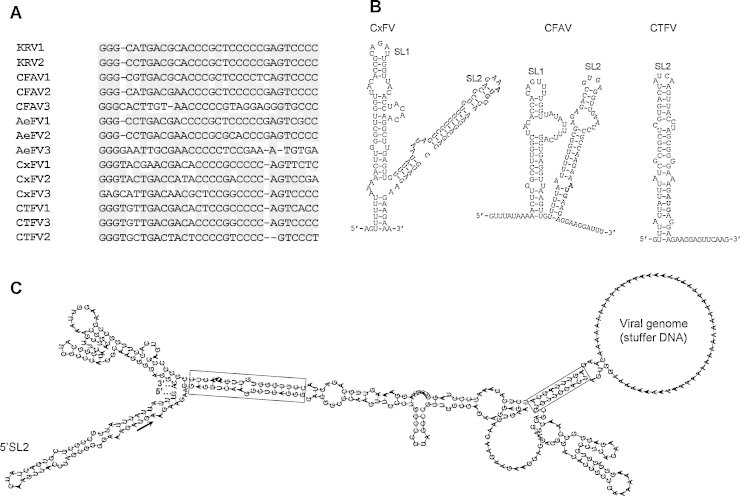
(A) Alignment of conserved GC-rich sequences located in the 3′-UTR of different insect-specific flavivirus (Aedes flavivirus – AeFV; cell fusing agent virus – CFAV; Kamiti River virus – KRV; Culex flavivirus – CxFV; Culex theileri flavivirus – CTFV). The most frequently found nucleotides at each position in the alignment are shaded in gray. (B) Predicted secondary structures for the Culex flavivirus – CxFV (AB262759), cell fusing agent virus (NC_001564) and Culex theileri flavivirus – CTFV (HE574574). SL indicates stem-loop structures. The AUG translation initiation codon is indicated in bold-face. (C) Computer-generated secondary structure analysis of possible interactions between the genomic plus-strand RNA ends of CTFV178 (HE574574). Sections of the 5′- and 3′-UTR are connected by a poly(A) insert (stuffer DNA) simulating most of the viral coding region. The AUG codon is indicated by the arrow. Regions of extended complementarity between both genome ends are boxed. The putative SL2 stem-loop that characterizes the 5′-UTR is indicated by 5′SL2.