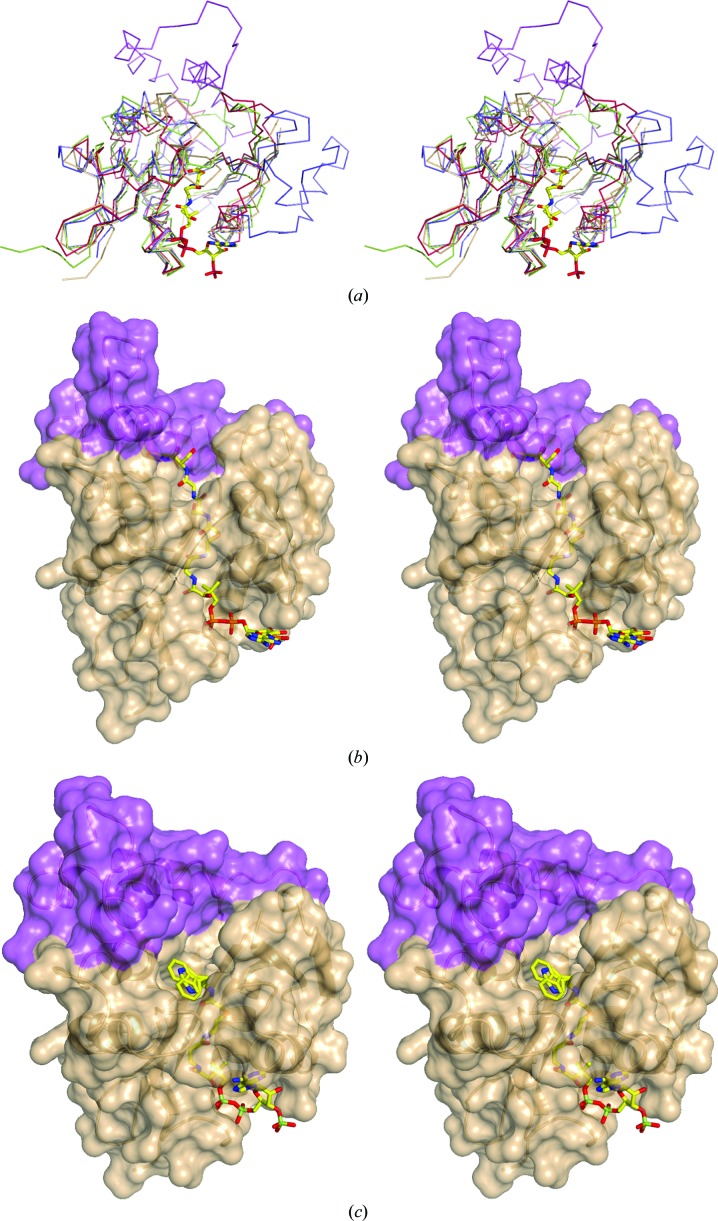
Figure 4.
Structural comparison of different acetyltransferases. (a) Superposition of the backbone of SsAT (fawn) with the 55-residue insertion (purple) with NATA (PDB entry 4kvm, dark red), serotonin N-acetyltransferase AANAT (PDB entry 1kuy, green) and aminoglycoside acetyltransferase AAC (PDB entry 2a4n, blue). The AcCoA moiety based on the NATA complex is shown in stick representation to mark the binding site. All structures show the same conserved core structure with the characteristic splay between β-strands 4 and 5 necessary for AcCoA binding. (b) Cartoon represention of SsAT with its solvent-accessible surface overlaid with the NATA bisubstrate complex (PDB entry 4kvm). The bound bisubstrate, shown in stick representation, binds with its AcCoA part in the binding site. The acceptor peptide moiety clashes with the unique 55-amino-acid insertion of SsAT (purple), which is not present in NATA. (c) The cartoon represention of SsAT with its solvent-accessible surface overlaid with the serotonin N-acetyltransferase bisubstrate complex (PDB entry 1kuy). The bisubstrate binds within the binding groove of SsAT with the acceptor tryptamine moiety pointing into a pocket and overlays partially with the remaining unmodelled density, attributed as a possible yet unidentified acceptor.