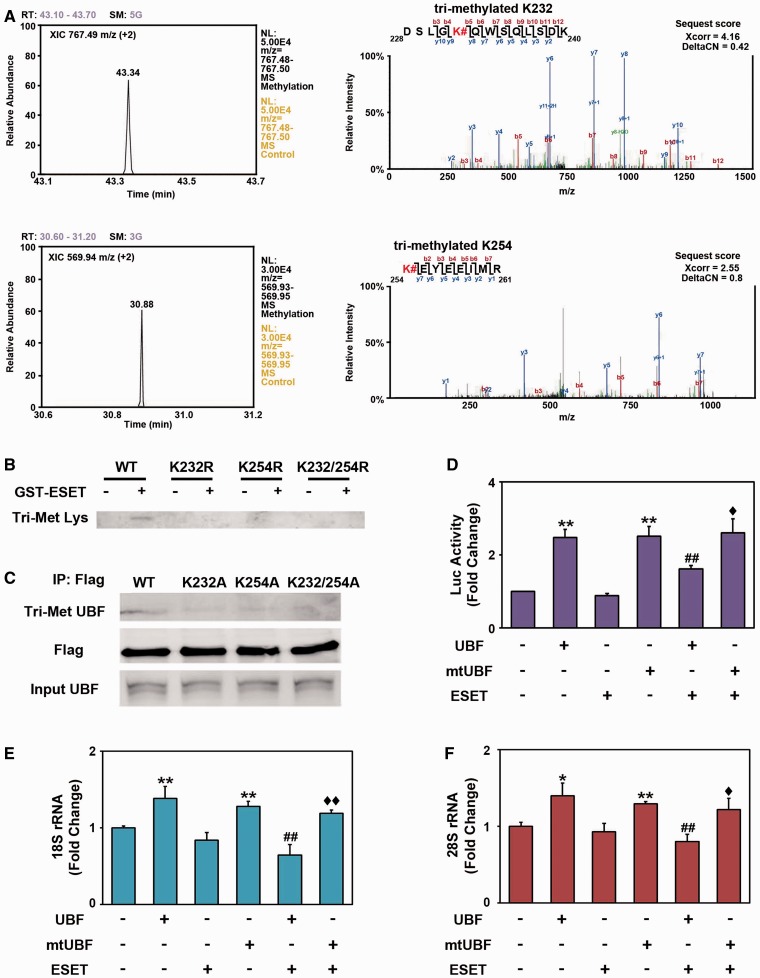
Figure 3.
UBF is methylated at K232/K254 residues by ESET and the methylation status of UBF contributes to the transcriptional activity of rDNA. (A) Liquid chromatography-tandem mass spectrometry analysis identified the methylation site of UBF at K232 and K254 by ESET. The XIC of the trimethylated K232 peptide (DSLGK#QWSQLSDK, m/z = 767.49, doubly change) and tri-methylated K254 peptide (K#EYEEIMR, m/z = 569.94, doubly change) were extracted within the narrow m/z range (m/z = 767.48 to 767.50 in DSLGK#QWSQLSDK and m/z = 569.93 to 569.95 in K#EYEEIMR) using Xcalibur software (Thermo). Representative MS/MS spectra of trimethylated peptides (DSLGK#QWSQLSDK and K#EYEEIMR) are shown in right panels. Fragment ions detected in our study are labeled. The methylation site is marked with a sharp. The SEQUEST matching scores (Xcorr and deltaCN) of peptides were also shown. (B) Mutations at K232R, K254R and K232/254R of GST–UBF–HMG2 domain abrogated the trimethylation by ESET in vitro. (C) Ectopic expression of UBF methylation site mutants (K232A, K254A and K232/254A) reduced the methylation of UBF in intact cells. pCMV-Flag-UBFs (WT, K232A, K254A and K232/254A) were transiently transfected, immunoprecipitated with anti-Flag antibody and blotted with anti-Tri-Met-Lys antibody. The whole blots of Tri-Met Lys in (B) and Tri-Met UBF in (C) are presented in the Supplementary Figure S8. (D) The methylation site mutant of UBF (K232/254A double mutant, DM) resulted in the recovery of rDNA transcriptional activity in response to ESET (n = 5). The methylation site mutant UBF [mtUBF (K232/K254A)] restored 18S (E) (n = 5) and 28S (F) (n = 3) rRNA levels that were decreased by ESET. Double asterisk represents significantly different from control at P < 0.005; Hash represents significantly different from UBF at P < 0.05; Diamond represents Ssignificantly different from UBF with ESET at P < 0.05.