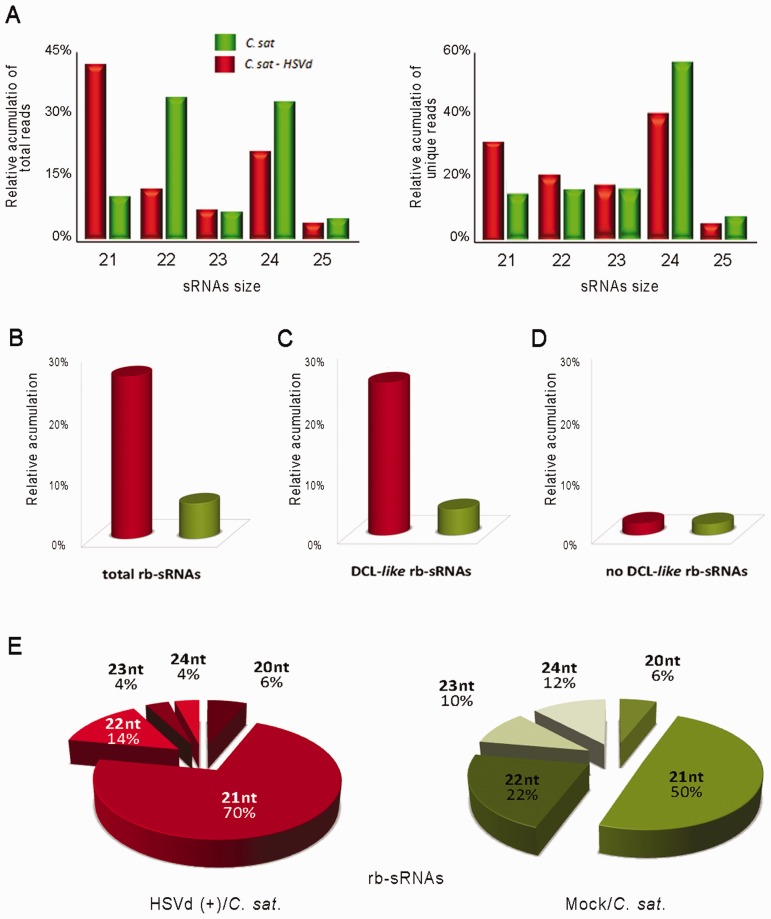
Figure 1.
Characterization of the sRNAs recovered from cucumber leaves by deep sequencing. (A) Graphic representation of the differential accumulation and distribution of the total (left) and unique (right) reads of endogenous cucumber sRNAs ranging between 21 and 25 nt recovered from both the control and HSVd-infected samples analyzed at 30 days post-inoculation. (B) Differential recovering of rb-sRNAs from infected and healthy tissues. The accumulation of rb-sRNAs is expressed as the percentage of total rb-sRNAs from the overall sRNAs in the library. (C) and (D) show the relative accumulation of recovered rb-sRNAs with canonical (21–24 nt) and non-canonical (<21 and >24 nt) predicted sizes, respectively. (E) Graphic representation of the distribution of the total reads of rb-sRNAs (ranging between 21 and 25 nt) recovered from both the HSVd-infected (left) and control (right) analyzed samples.