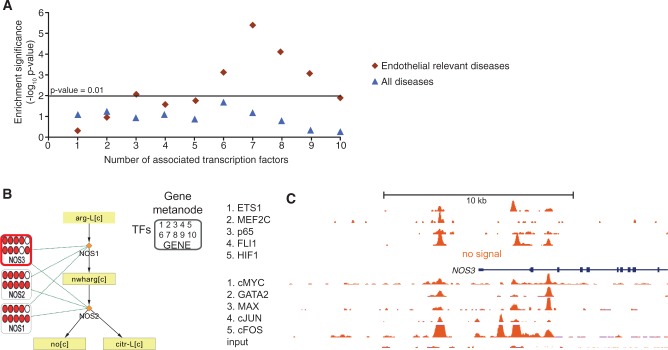
Figure 2.
Gene metanodes reveal frequent TF association to disease-relevant genes exemplified by nitric oxide synthases in HUVEC data. The result of disease enrichment tests for genes associated with 1–10 TFs are shown (A), where the horizontal line corresponds to hypergeometric P <0.01. The endothelial relevant diseases (diamonds) is compared with all diseases (triangles). (B) The three enzymes encoding nitric oxide synthases (NOS1, NOS2 and NOS3) and the two reactions that convert l-arginine to NO for regulating vascular dilation are shown. Data related to genes are displayed in gene metanodes superimposed on the metabolite-reaction network. Peak associations from 10 ChIP-seq studies in HUVECs (ETS1, MEF2C, p65, FLI1, HIF1α available via NCBI SRA and cMYC, GATA2, MAX, cJUN, cFOS and input available via ENCODE) are displayed in the indicated order where color indicates TF association and the respective TF signal tracks are shown in C. The value range 1–100 is used in the first five tracks and 1–78 in the ENCODE tracks.