Figure 1.
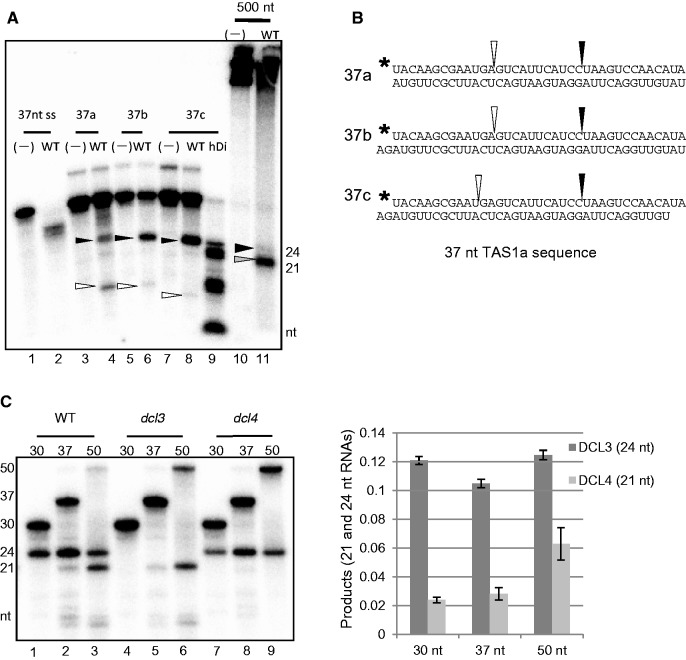
DCL3 and DCL4 preferentially cleave short and long dsRNAs, respectively. (A) DCL3 efficiently cleaves short dsRNAs 37 nt long. For a Dicer assay, 32P-labeled dsRNAs of 37 and 500 nt were incubated with crude extracts from WT Arabidopsis seedlings for 2 h at 22°C. Cleavage products were analysed by 15% denaturing PAGE. Abbreviations: 37 nt ss, 37 nt single-stranded RNA; 37 a, 37 b and 37 c, 37 nt dsRNAs with various ends indicated in (B). 37 nt ssRNA and dsRNAs were phosphorylated at the 5′ end by [γ-32P]-ATP. 500 nt, 500 nt dsRNA whole-labeled by 32P-UTP. (–), no crude extracts (negative control). A recombinant human Dicer fragment (hDi) was used as a cleavage control to generate size markers. Black (24 nt) and white (11 or 13 nt) arrowheads indicate cleavage products produced by DCL3, and a gray arrowhead (21 nt) indicates cleavage products by DCL4. (B) Structures of substrate dsRNAs of 37 nt, which have a TAS1a gene sequence, and cleavage sites (black and white arrowheads) by DCL3. Black stars indicate 32P. (C) DCL3 cleaves short dsRNAs, whereas DCL4 cleaves long dsRNAs. 5′-32P-phosphorylated dsRNAs of 30, 37 and 50 nt with 2 nt 3′ overhangs were incubated with crude extracts from WT, dcl3-1 or dcl4-2 Arabidopsis seedlings for 2 h at 22°C. Autoradiography (left) results are shown. The 21 nt RNA producing activity (DCL4 activity) was calculated as the relative band intensity of 21 nt RNA with respect to the intensity of all bands including smeary bands in each lane, and the 24 nt RNA producing activity (DCL3 activity) was calculated using the same equation. Values are means ± standard deviation for three independent experiments (right).