Figure 3.
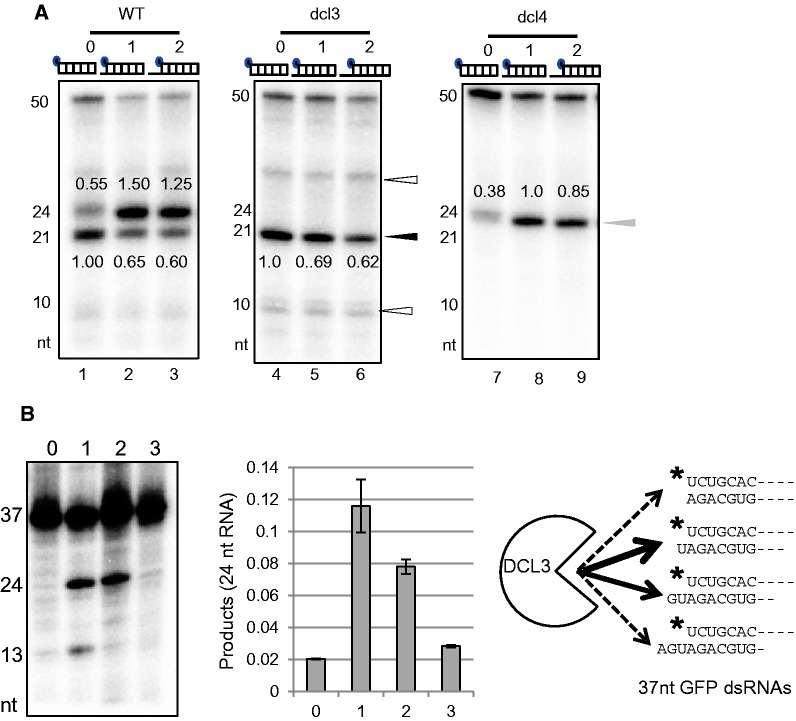
DCL3 cleaves dsRNAs with 1 nt 3′ overhang more efficiently than dsRNAs with 2 nt 3′ overhang or a blunt end, but DCL4 cleaves these dsRNAs with similar efficiency. (A) Cleavage products of three 50 nt dsRNAs with various 3′ ends formed by DCL3 or DCL4 and analysed by denaturing PAGE. Three 50 nt dsRNAs, which had blunt ends or 1 or 2 nt 3′ overhangs and contained a TAS1a gene sequence, were used as substrates for Dicer assays with WT, dcl3-1 or dcl4-2 crude extracts. Amounts of cleavage products, 21 nt RNA (DCL4) and 24 nt RNA (DCL3), were calculated as the relative band intensities of 21 and 24 nt RNAs, respectively, with respect to the intensity of all bands in each lane following autoradiography. Values in each lane are relative signal intensities of cleavage products, defining the signal intensity of 21 or 24 nt product in lane 1, 4 or 8 as 1.00. A black arrowhead indicates 21 nt cleavage products obtained from the 5′ phosphorylated end by DCL4, and white arrowheads indicate ∼10 and 30 nt cleavage products from the non-phosphorylated end by DCL4. A gray arrowhead indicates 24 nt cleavage products from the 5′ phosphorylated end by DCL3. (B) DCL3 preferentially cleaves dsRNAs with a 1 nt 3′ overhang. Four 37 nt dsRNAs, which had blunt ends or 1–3 nt 3′ overhangs and contained a GFP gene sequence, were used as substrates for Dicer assays. The 24 nt RNA producing activity (DCL3 activity) was calculated as the relative band intensity of 24 nt RNA with respect to the intensity of all bands in each lane following autoradiography. Values are means ± standard deviation for three independent experiments. The schematic figure shows cleavage preference of DCL3. Solid arrows indicate cleavage preference of DCL3. Black stars indicate 32P.